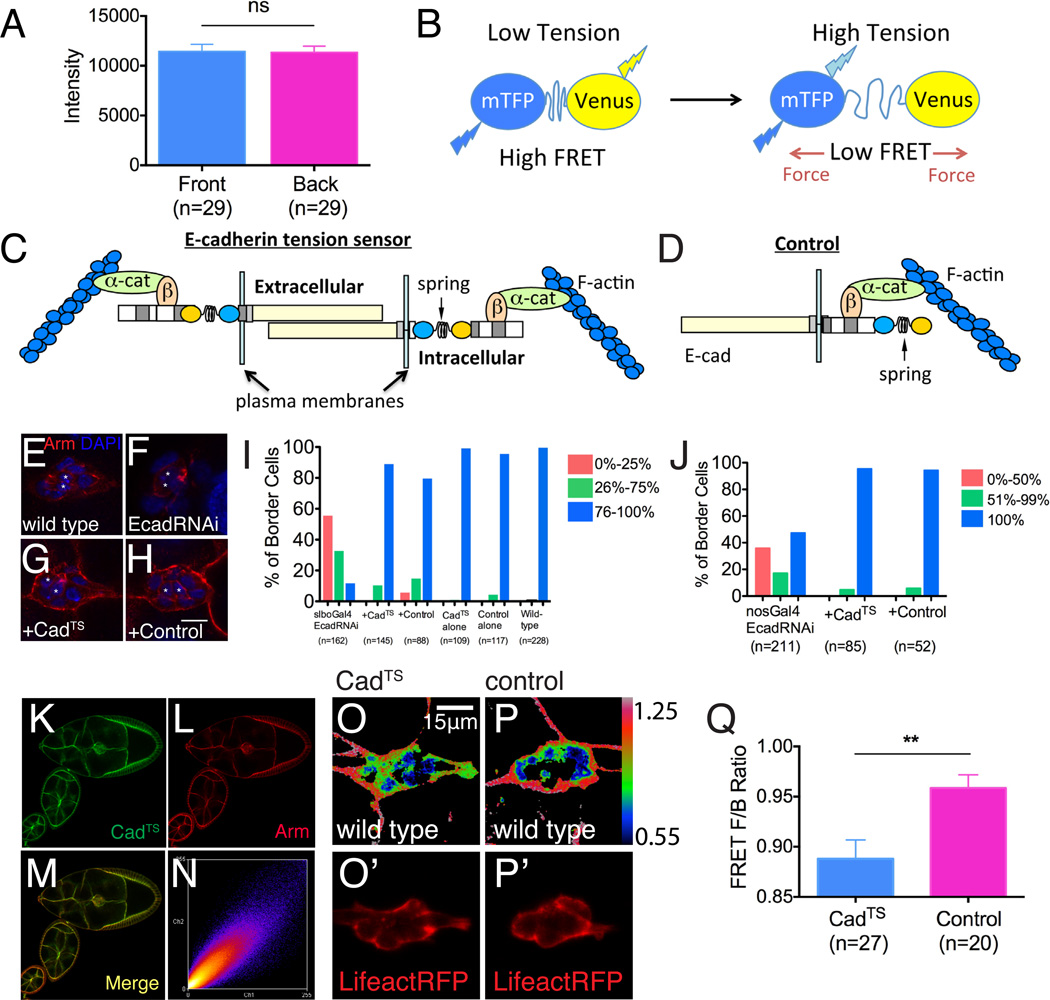
Figure 3. An in vivo E-cadherin tension sensor.
(A) Quantification of E-cadherin immunofluorescence intensity at the front and back of migrating border cell clusters. Data are presented as mean ± SEM. (B) Schematic drawing of the tension-sensing (TS) module. Teal fluorescent protein (mTFP) is separated from Venus, a yellow fluorescent protein, by a nano-spring protein domain from spider silk. In the relaxed state, the two fluorophores are close enough to allow FRET. The spider silk domain stretches in response to pico Newton forces, reducing FRET (Grashoff et al., 2010 see text and Experimental Procedures for details). (C) Schematic of the E-cadherin tension sensor (CadTS), and (D) a corresponding control construct, which should not be tension-sensitive. (E-H) Rescue of Armadillo expression (Arm, which is Drosophila β-catenin) in border cells after EcadRNAi (F) by CadTS (G) and control (H). Polar cells are marked by asterisks. Scale bar shows 10 μm. (I-J) Histograms showing CadTS and control rescuing border cell migration after border cells-specific (I) and nurse cell-specific (J) EcadRNAi. (K-N) Colocalization of CadTS with Arm. (O-P’) FRET images of border cell CadTS (O) and control (P) pseudo-colored in Rainbow RGB. The outlines of border cells are shown by Lifeact-RFP which is co-expressed in the same experiment (O’, P’). (Q) Histogram showing the front to back FRET ratios for CadTS (blue) and control (pink). Data are presented as mean ± SEM. **p<0.005. See also Supplemental Figure S2.