Abstract
Copy number variation (CNV) of human chromosome 22q11.2 is associated with an elevated rate of autism spectrum disorder (ASD) and represents one of syndromic ASDs with rare genetic variants. However, the precise genetic basis of this association remains unclear due to its relatively large hemizygous and duplication region, including more than 30 genes. Previous studies using genetic mouse models suggested that although not all 22q11.2 genes contribute to ASD symptomatology, more than one 22q11.2 genes have distinct phenotypic targets for ASD symptoms. Our data show that deficiency of the two 22q11.2 genesTbx1 and Sept5 causes distinct phenotypic sets of ASD symptoms.
Keywords: Tbx1, Sept5, 22q11.2, Syndromic ASD, Copy number variation
Introduction
Genes are currently the best available entry point for the studies aimed at understanding the brain mechanisms underlying autism spectrum disorders (ASD). Early twin studies of ASD indicated the proportion attributable to genetic factors at about 90% [1–5]. Although a more recent, large-scale study with recent ASD criteria has estimated a lower rate of ASD heritability [6], it is still safe to conclude that genetic variation confers a considerable risk for ASD.
In an attempt to identify individual contributory genes, various types of genetic variants are being examined. Genome-wide association with many single nucleotide polymorphisms (SNPs) suggest that commonly found variants confer a 1.2–3 fold increase in risk for ASD [7]. Additionally, rare and genetically identifiable cases of ASD or syndromic ASDs are being explored. They include mutations of single genes and copy number variations (CNVs). While it remains unclear if rare variants and common variants seen in ASD share alterations in similar or overlapping molecular cascades and networks, rare variants are often associated with substantially increased risk for ASD than common variants, and thus study of rare variants is the best currently available approach towards identification of ASD mechanisms.
22q11.2 CNV represents a Syndromic ASD
Our group has focused on human chromosome 22q11.2 as a reliable genetic risk factor for ASD. Deficits in social behavior, skills and cognition have long been noted in 22q11.2 hemizygous children [8–15]. Fourteen to 50% of individuals with 22q11.2 hemizygosity examined for ASD are reported to meet diagnostic criteria [12,16–21]. Patients with 22q11.2 duplication meet criteria for ASD when evaluated using the Autism Diagnostic Observation Scale (ADOS), Autism Behavior Checklist (ABC), and Childhood Autism Rating Scale (CARS) [22–25]. However, patients with 22q11.2 CNV are often referred for formal psychiatric evaluation only after they exhibit cognitive, social and behavioral problems (i.e., ascertainment bias). Moreover, the number of duplication cases so far identified is not large enough to permit computation of the true rate of ASD. Nevertheless, when screened from the general ASD population, 22q11.2 hemizygosity and duplications have been identified as rare variants in many studies [26–34].
22q11.2 CNV and other Neuropsychiatric Disorders
Individuals with 22q11.2 hemizygosity exhibit other neuropsychiatric disorders, including severe, mild and borderline mental retardation (50–90%) [12,13,35–39], attention-deficit/hyperactivity disorder (35–55%) [12,38,40–44], obsessive compulsive disorder (8–33%) [41–45], schizophrenia (~25%) [35,43,45–52], generalized anxiety disorders (10–28.6%) [12,42,43], schizoaffective disorder (2–8%) [41,44,45,48], and other behavioral problems as well as phobias and anxiety disorders [12,40–45].
Karayiorgou and colleagues [53] pointed out that ASD and most diagnoses noted above, except schizophrenia, might not be genuinely associated with 22q11.2 hemizygosity. It is true that high rates (~25%) [50] of schizophrenia are associated with 22q11.2 hemizygosity [35,43, 45,47,48,54]. Clearly, more evidence is needed to associate 22q11.2 hemizygosity with additional diagnoses. However, ASD diagnosis was made by experienced raters and psychiatrists based on validated and reliable scales, such as the Autism Diagnostic Interview—Revised (ADI-R) and Diagnostic and Statistical Manual of Mental Disorders-IV (DSM-IV), in studies that reported higher than expected rates of ASD [16–20]. It is premature to dismiss the notion that heightened rates of ASD also are associated with 22q11.2 CNV.
While some subtle differences have been noted in symptomatic elements between a small sample of children with 22q11.2 hemizygosity and idiopathic autistic children [19], it is unclear if such subtle differences in a small sample size invalidate the ASD diagnosis given the generally variable nature of symptomatic presentation in idiopathic ASD. Similarly, Eliez [55] reported that children with hemizygosity exhibit language impairment but catch up following surgical and therapeutic interventions of cleft palate and their verbal reasoning skills are stronger than those for nonverbal reasoning; idiopathic autism is associated with weaker verbal profiles compared to nonverbal profiles throughout development. Only 10 of 300 children exhibited impaired verbal abilities among Eliez’s sample with 22q11 hemizygosity. However, language impairments are variable in 22q11.2 hemizygous babies and children[55]; although a majority show lower performance IQ than verbal IQ, a sizable subpopulation shows the reverse pattern [56]. Moreover, as children with 22q11.2 grow, verbal IQ declines more rapidly than performance IQ and verbal IQ becomes lower than or comparable to performance IQ [54, 57]. Idiopathic ASD children also have varying degrees of language delays [58, 59] and importantly, many of those with language delays become fluent speakers by later school years [60].
It has also been suggested that underlying processes (e.g., social motivation and social skills) may be different between 22q11.2 associated and idiopathic ASDs. Although individuals with idiopathic ASD are impaired in both motivation for social interaction with others [61–63] and processing of social cues and understanding the mental state of others (known as theory of mind) [64,65], a high degree of heterogeneity is noted and in fact, some display genuine signs of social motivation but lack the skills [61]. Thus, these processes do not provide a clear-cut discriminating power to differentiate between idiopathic and 22q11.2-associated ASD.
Although most of 22q11.2-associated neuropsychiatric disorders are not found at a higher frequency among individuals with the 22q11.2 microdeletion than in cohorts with other developmental disorders associated with learning disabilities [53], it should be noted that in the idiopathic ASD population, ASDs are associated with high rates of comorbidity with severe cognitive impairments [66–69] and intellectual disabilities [70]. Similarly, individuals with 22q11.2-associated ASD have high rates of developmental delays and cognitive impairments [12,13,35–39,71,72]. Given this comorbidity, it is not certain if there is a specific brain development and functional mechanism that is so selectively affected that only ASD is manifested without comorbidity.
It is true that a significant enrichment for 22q11.2 deletions was not found in ASD samples in some studies [53]. Ogilvie and colleagues reported no case with 22q11.2 deletion among 103 ASD patients from multiplex families [73], but this sample size is not sufficient for detection of a rare CNV. In another study of simplex and multiplex ASD cases, 22q11.2 duplications, but not hemizygosity, were enriched [74]. However, many other studies reported enrichment for 22q11.2 duplications and hemizygosity in ASD samples [26–33], and a combined analysis of studies with stringent criteria demonstrated statistically significant enrichment of 22q11.2 CNV in 3,816 ASD samples [34]. Statistically significant enrichment of any rare CNV is generally difficult to achieve after correction for multiple comparisons, due to its very rare nature [34]. Detection of 22q11.2 hemizygosity in ASD samples in simplex and multiplex cases is additionally complicated by the relatively higher rates of de novo as opposed to inherited hemizygosity [75–78] and the opposite trend for duplications [79–82].
It was suggested that diagnoses of ASD might reflect misdiagnosis of social impairments actually associated with premorbidity in schizophrenia [53]. Eliez [55] reported that 56% of children with childhood-onset schizophrenia are first diagnosed with pervasive development disorder (PDD), while rates for diagnosis of autism during childhood and schizophrenia later in life are less than 5%. However, one retrospective analysis indicates that half of schizophrenic patients meet the genuine diagnostic criteria for ASD during childhood [83]. More work is needed to dismiss the possibility that 22q11.2 hemizygosity increases susceptibility to both schizophrenia and ASD.
Mouse Models of 22q11.2 CNV
It has not been feasible to ascertain the impact of dose alterations of individual 22q11.2 genes within the 1.5–6Mb CNV region on various phenotypes in humans. Association of single nucleotide polymorphisms (SNPs) on the remaining copy of 22q11.2 in individuals with ASD determines how such alleles modify phenotypes of 22q11.2 hemizygosity, but does not identify genes whose hemizygosity causes phenotypes. Moreover, SNPs are not equivalent to deletions or duplications and do not consistently confer susceptibility to neuropsychiatric disorders [84].
Modeling genetic abnormalities of 22q11.2 CNVs is relatively straightforward due to conserved sequence homology between the mouse and human. The usefulness of a rodent model resides in its ability to precisely manipulate a specific gene in isolation and predict its outcome; this is not possible in humans because human studies are, in essence, observation of correlation. We and others have used genetically engineered mouse models to identify small segments and single 22q11.2 genes responsible for ASD-related behavioral phenotypes (Figure 1).
Figure 1. Genetic mouse models of 22q11.2 CNVs.
Over-expression (left) and deletion (right) cases are indicated. Vertical bars indicate the extent of chromosomal segments over-expressed or deleted. Phenotypes consistent (red) and inconsistent (black) with those associated with ASD are shown. a) hyperactivity, sensitization, social behaviors and clozapine-response are measured [85]; b) social interaction, working memory, prepulse inhibition (PPI), and anxiety and motor behavior were measured [93]; c,d,e,f and g) auditory PPI was measured. c[97], d[98], e[99], f[100], and g[94].
It is inherently difficult to behaviorally model ASD symptoms in mice and any attempt to model symptoms in experimental animals is at best a proxy for the real behaviors/symptoms. While modeling overall symptomatology is difficult, ASD may be more reliably characterized when a link is sought between genetic risk factors and dimensions of a specific behavioral element of ASD. We have measured specific behavioral elements of ASD, including social interaction, social communication and repetitive behavior. Ultimate validation of the efficacy of mouse models will only be accepted when hypothetical mechanisms of ASD and therapeutic effectiveness in an animal model are consistent with observations in humans.
What has emerged from these mouse studies is the knowledge that not all 22q11.2 genes contribute to ASD-related behavioral phenotypes. In 2005, our group reported that mice over-expressing a ~200 kb segment of human 22q11.2, containing Gnb1l, Tbx1, Gp1Bβ and Sept5, exhibit hyperactivity, spontaneous sensitization, lack of normal social interaction (Figure 1a; see also Supporting Information, Movie 2 in [85]). Spontaneous sensitization of hyperactivity was completely blocked after three weeks of treatment with the antipsychotic drug clozapine [85]; clozapine and related atypical antipsychotic drugs attenuate some ASD symptomatic elements [86]. These phenotypes were present as early as 5 weeks old and persisted up to 2–4 months of age. However, the level of hyperactivity in this mouse model was so high that it might have rendered mice physically unable to engage in reciprocal social interaction. It was not technically possible to analyze more detailed affective and cognitive behaviors due to the extraordinarily high levels of hyperactivity.
We subsequently demonstrated that over-expression of an adjacent ~190 kb segment, containing Arvcf, Comt and Txnrd2, impaired working memory (consistent with deficits seen in 22q11.2 hemizygous patients [11,87–91] and idiopathic ASD patients [92]), but had no effect on reciprocal social interaction or prepulse inhibition (PPI) [93] (Figure 1b). However, working memory has not been examined in 22q11.2 duplication patients so far, and relevance of this mouse phenotype to duplication phenotypes remains unclear. The fact that this chromosomal segment dissociated working memory from PPI and social interaction suggests that these behavioral phenotypes are genetically dissociable.
Stark and colleagues provided complementary evidence that over-expression of chromosomal segments outside the 200 kb region does not induce PPI deficits [94]. Mice overexpressing a segment containing Prodh and Vpreb2 exhibited a higher level of PPI than WT mice (Figure 1g). It is not clear whether this mouse phenotype is consistent with that in humans, because, to date, PPI has not been examined in duplication cases. Moreover, given that both 22q11.2 duplication and hemizygosity are associated with ASD, it might be expected that high and low doses of 22q11.2 cause phenotypes in the same, not opposite, direction. A second mouse line had over-expression of a segment that included Zdhhc8, Ranbp1, Htf9c, T10, Arvcf and Comt (Figure 1g); this mouse was indistinguishable in PPI from WT mice. This was consistent with our own data showing that the 190 kb transgenic mouse over-expressing COMT and two other genes showed normal PPI (Figure 1b) [93]. Similarly, Weinberger’s group demonstrated that Comt over-expression or deletion does not affect PPI [95]. Given that Comt elevation nevertheless impairs working memory in these mice [93,95], elevated levels of this 22q11.2 gene seem to selectively impair working memory without impacting PPI [93,95] or social interaction [93].
Taken together, these observations suggested that the 200 kb segment we identified (Figure 1a) [85] might contain a gene or genes that contribute to behavioral phenotypes related to ASD. The fact that over-expression of the 200 kb region alone was sufficient to induce behavioral phenotypes related to ASD is of considerable interest, as it implies that this genomic abnormality could act as a primary causative event rather than a susceptibility factor. Note that the phenotypic targets of individual 22q11.2 genes are not identical. Over-expression of the 200 kb region causes a number of behavioral phenotypes related to ASD, whereas that of the adjacent 190 kb region results in selectively impaired working memory.
Children with 22q11.2 hemizygosity exhibit defective auditory PPI [96]. The genetic origin of this behavioral phenotype was identified by a series of elegant mouse studies. Several groups examined the effects on PPI of 1.5 Mb or smaller, partly overlapping deletions of murine chromosome 16, a mouse ortholog of human 22q11.2 (Figure 1). Auditory PPI was defective only when large deletions encompassed the same 200 kb region; when large deletions occurred outside the 200 kb region, no PPI deficit was seen [97–100] (Figure 1c,d,e and f). These reports conclusively demonstrated that the same 200 kb region is also responsible for this behavioral phenotype in 22q11.2 hemizygosity.
Collectively, these mouse studies form a solid basis upon which to further study genetic mechanisms of 22q11.2-associated ASD. Our subsequent studies have focused on two genes encoded in the 200 kb region in mouse models.
Tbx1
A rare case of TBX1 mutation (not 22q11.2 hemizygosity) was associated with Asperger syndrome in one individual [97]. Tbx1 is one of four genes encoded in the 200 kb region and belongs to a phylogenetically conserved family of genes that share a common DNA-binding domain, the T-box. The human TBX1 protein and its mouse ortholog Tbx1 share a highly conserved amino acid sequence. Tbx1 mRNA is present at low levels in the embryonic mouse brain and is expressed at increasingly higher levels in the postnatal and adult mouse brain [97].
Reverse transcription-polymerase chain reaction (RT-PCR) analysis showed Tbx1 mRNA expression in the prefrontal cortex, nucleus accumbens, caudate-putamen, amygdala, hippocampus, ventral tegmental area, and substantia nigra of C57BL/6J mice at 2 months of age [101]. Immunofluorescent analysis similarly showed that low signal levels of Tbx1 were present in many brain regions of 2 month-old C57BL/6J mice, but higher levels were found in the rostral migratory stream, the dentate gyrus, and the subventricular zone. These data are consistent with the reports that Tbx1 mRNA and protein are present in the whole adult mouse brain samples [97,102], and further reveal the presence of Tbx1 mRNA and protein in distinct brain regions. Interestingly, these brain regions are known to undergo postnatal and adult neurogenesis. In fact, higher Tbx1 protein levels have been reported during proliferation than differentiation in neural progenitor cell cultures derived from the hippocampal dentate gyrus [101].
Note that Tbx1 has been deposited as an alias of mouse lipopolysaccharide-induced TNF factor (Litaf) at one NCBI site (GenBank: AF171100.1; http://www.ncbi.nlm.nih.gov/nuccore/AF171100) despite the fact that these two genes have different sequences and different chromosomal locations (Tbx1, Mus musculus chromosome 16, 18581713-18586969; Litaf, Mus musculus chromosome 16, 10959273-10993121). This error has propagated other Tbx1 and Litaf listings on the NCBI, MGI and many other similar sites and might be a reason why one published comprehensive analysis of 22q11 gene expression used “Tbx1” primers that have no sequence homology with Tbx1 and reported that “Tbx1” mRNA signals, which are in reality litaf signals, were not detectable in any brain regions of adult mice.
Although, we noted sensitized hyperactivity in 200 kb transgenic mice (Figure 1a) [85], relevance of this behavior to ASD is also not clear. While clozapine attenuated hyperactivity is caused by over-expression of the 200 kb [85] segment and it is known that this drug attenuates some symptoms of ASD [86], it is unclear whether sensitized hyperactivity in mice models the core symptoms of ASD.
While PPI is a reliable parameter for sensorimotor gating [103], its relevance to ASD has not been definitively established. Defective PPI is not consistently seen in individuals with ASD [104–107]. Moreover, evidence suggests that PPI, as an endophenotype, is genetically dissociable from symptomatic elements of ASD and schizophrenia. For example, in Sept5 KO mice, social interaction is reduced but PPI is potentiated [108–110].
We, thus, examined social interaction in a naturalistic social interaction paradigm in which an age-matched, male C57BL/6J inbred mouse was paired with either a congenic Tbx1 HT mouse or WT mouse; Tbx1 homozygous mice are not viable. As a pair of mice is placed in a cage that is novel to both, there is no ‘resident’ mouse in this task; aggressive social interaction is minimized and affiliative social interaction is maximally evaluated [93,108]. Unlike a “sociability” task in which one of the mice is confined in a small wire cage, reciprocal interaction can be evaluated in this naturalistic social interaction task [111]. Tbx1 HT mice exhibited significantly lower levels of active and passive affiliative social interaction (Figure 2A); no detectible aggressive social behavior was seen in this setup.
Figure 2. A) Active affiliative (affiliative), aggressive, and passive affiliative (pasasive) forms of social interaction inTbx1 WT and HT mice.

Time spent(mean ± SEM) in the three forms of social interaction in two 5-min sessions with an age-matched stimulus C57BL/6J mouse is shown. Asterisks indicate statistically significant differences between WT and HT mice at levels of 0.05(*) and 0.01(**), as determined by Newman-Keuls comparisons. B) Ultrasonic vocalization of pups during a 5-min separation from mothers at postnatal days 7–8. The average duration (mean±SEM) of each vocal call type is shown. Distinct catagories of calls, as defined by Scattoni and colleagues [138], are indicated as: cx, complex; ham, harmonics; ts, two syllable; u, upward; d, downward; h, hump(a.k.a., chevron); sh, shorts; c, composite; fs, frequency steps; f, flat. An asterisk indicates a statistically significant difference between WT and HT mice at levels of 0.05(*) and 0.01 (**) as determined by Newman-Keuls comparisions. C) Spontaneous alternation in T-maze. The percentage of repeated visits to the same am (mean ± SEM) is shown. Mice were tested with 0-, 30-, and 60-s delays between trails. An asterisk indicates a statistically significant difference between WT and HT mice at 0.01 (**) at each delay (solid line), as determined by Newman-Keuls comparisions. This figure is reproduced from [101] with permission of the Oxford Press.
Babies and children with 22q11.2 hemizygosity exhibit delayed development of vocal volume, vocalization, and language [112] and social communication deficits [8–21]. When mouse pups are separated from mothers, they typically emit ultrasonic vocalization. This vocalization elicits their retrieval by mothers, and thus is considered a form of social communication in rodents [113,137]. We examined ultrasonic vocalization at postnatal days 7–8. HT mice exhibited vocalization for shorter duration in harmonic, two-syllable, composite, and frequency steps, compared to WT littermates (Figure 2B). Interestingly, these defective vocalization patterns in HT mice are fairly complex, but WT and HT vocalizations were indistinguishable in simple patterns (e.g., upward, downward, hump, and short).
Mice have a natural tendency to alternate arms visited in a T-maze, a behavior that requires working memory to recall a previously arm visited and alternate visits [114]. Tbx1 HT mice showed higher levels of repeated visits to the same arm (Figure 2C). When HT mice showed working memory at 0 seconds delay, they had a higher degree of repetitive choices than WT mice; at a 60-sec delay, HT and WT mice were indistinguishable in repetitiveness and HT mice did not show increased repetitiveness beyond 50% (Figure 2C). These data suggest that the repetitive behavioral tendency is present in HT mice only when it depends on working memory, and is not indicative of simple motor repetitiveness. It is interesting to note that individuals with idiopathic ASD have difficulty in inhibiting context-inappropriate behavior based on working memory; this is thought to underlie actions and verbalizations that are inappropriate in terms of timing or appropriateness to the circumstances; they are not impaired in simple response inhibition that is not dependent on memory [115,116]. Taken together, Tbx1 heterozygosity recapitulates symptomatic features of 22q11.2-associated ASD.
Sept5
Sept5 is abundantly expressed in rodent and human brains [117,118], and is presynaptically located to regulate neurotransmitter release at synapses together with the SNARE complex [119–121]. This protein additionally contributes to the structural health of axons and dendrites [122,123]. Given that synaptic alterations in synaptic and neuronal connection are seen in many mouse models of ASD, and our results showed that a gene dose alteration of the 200 kb region, including Sept5, impaired social interaction [85], we evaluated two issues regarding the functional role of Sept5 in ASD-related behavioral phenotypes.
Firstly, since not all individuals with 22q11.2 hemizygosity show ASD (i.e., incomplete penetrance) [12,16, 21], we hypothesized that genetic background affects phenotypic expression of Sept5 deficiency. Second, while limbic region activation occurs when humans are exposed to social cues and this activation is altered in individuals with ASD, the genuine functional role of these alterations in ASD and the brain regions through which Sept5 functionally mediates social behavior are not known. We hypothesized that Sept5 levels in the two major limbic regions (hippocampus and amygdala) are a determinant of social interaction.
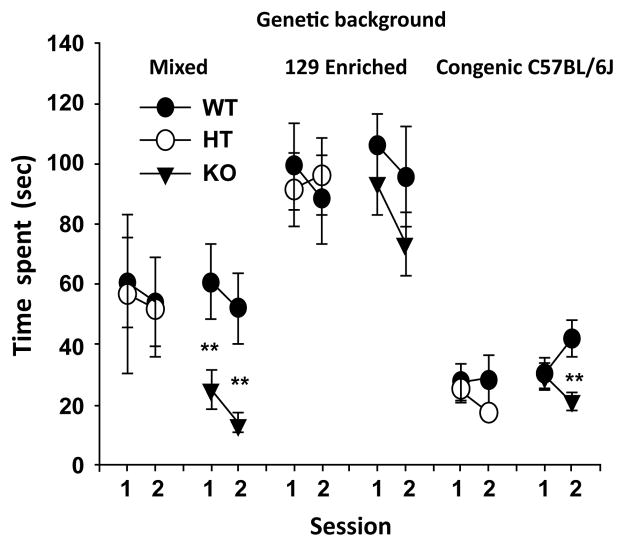
To address the first issue, we tested the impact of Sept5 deficiency on social interaction on three genetic backgrounds. Active affiliative social interaction was impaired in Sept5 homozygous (KO) mice with a mixed genetic background of CD1, 129X1/SvJ and 129S1/Sv-p+ Tyr+ Kitl SI-J/+ (Figure 3, Mixed) and with a congenic background with C57BL/6J (Figure 3, Congenic C57BL/6J), but not with a 129S1-enriched genetic background (Figure 3, 129 Enriched) [108]. Sept5 KO mice were not impaired in other behavioral measures, including working memory and repetitive behavioral trait (spontaneous alternation), PPI, anxiety-related traits, and motor activity [108–110], underscoring a rather selective action of Sept5 deficiency on symptomatic elements of ASD. Given that Sept5 deletion is included in 22q11.2 hemizygosity in humans, this gene is likely to contribute to one symptomatic element of ASD. A corollary of this observation is that as long as a gene deficiency causes at least one (not necessarily all) aspect of ASD in a mouse model, a gene should be considered to be a contributory one.
Figure 3.
Impact of genetic background on active social interaction in Sept5 deficient mice. Interaction time (mean ± SEM) spent in active social interaction between mice is shown in two successive 5-min sessions. Asterisks indicate statistically significant differences from WT mice at 1% (**) levels, as determined by Newman-Keuls comparisions. This figure is reproduced from [108,109] with permission of the Oxford Press.
Interestingly, Sept5 heterozygous mice were not impaired in affiliative social interaction, while 22q11.2 hemizygosity is sufficient to induce a high rate of ASD in humans. However, it is not known whether a gene-dose manifests itself similarly in mice and humans. Moreover, Sept5 heterozygosity is a single gene deficiency, but 22q11.2 hemizygosity carries deficiencies of multiple genes and heterozygosity of other 22q11.2 genes might amplify the impact of Sept5 heterozygosity in humans.
Our finding offered a plausible explanation for incomplete penetrance, but it did not entirely rule out the possibility that the phenotypic difference between congenic Sept5 WT and KO mice is caused by allelic heterozygosity instead of –or possibly in addition to—Sept5 deficiency. A simple estimate based on the number of backcrossings is that our congenic WT and KO mice are homozygous with C57BL/6J alleles at up to 99.8% of loci and the remaining fraction is heterozygous for alleles. However, one often ignored caveat of this estimate is that allelic homozygosity greatly differs at loci linked compared to those not linked to the gene of interest [124]. Even after 10 generations of back-crossing, more alleles from 129 mice are expected to be present at loci near the Sept5 gene in KO mice than in WT mice. Thus, our observation still does not rule out the possibility that social interaction deficits in congenic KO mice reflect allelic differences rather than Sept5 deletion. Currently available breeding techniques do not offer a definitive technical option to entirely rule out this possibility [125,126]. There are many mouse models of ASD with non-congenic background. Caution is needed in ascribing a phenotypic difference between mutant and wild type mice to the impact of the mutant gene rather than allelic differences in the genetic background. It would be interesting to observe how the phenotypic expression of other ASD-related genes is modified by genetic background in other mouse models of ASD.
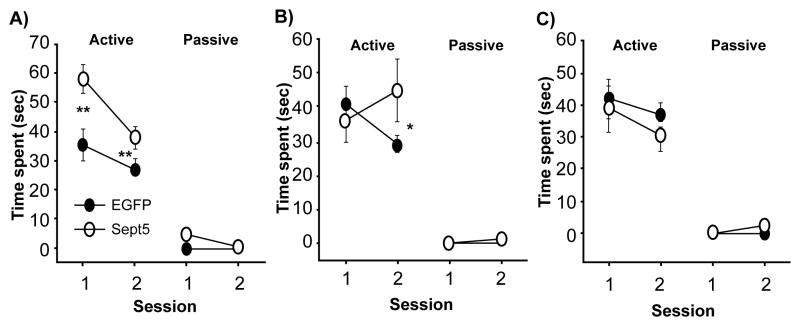
To address this interpretative caveat and identify brain regions in which Sept5 levels regulate social interaction, we expressed Sept5 in selected brain regions at the time of behavioral testing in inbred C57BL/6J mice [109]. We constructed a lentiviral vector carrying Sept5 and surgically infused it into the brains of C57BL/6J mice, thereby elevating only Sept5 in distinct brain regions in a congenic genetic background. Compared to control mice that received enhanced green fluorescent protein (EGFP) alone, C57BL/6J mice that received Sept5-EGFP over expression in the hippocampus (Figure 4A) and amygdala (Figure 4B) showed increased active, affiliative social interaction. This phenotype was highly selective; Sept5 overexpression had no effect on reaction to a novel, non-mouse object, olfactory senses, anxiety-related behaviors or motor behavior [109]. Moreover, Sept5 over-expression in the somatosensory cortex had no effect on social interaction (Figure 4C). Although synaptic alterations in the sensorimotor cortex have been observed in some mutant mouse models of ASD and cortical development has been suggested to be aberrant in ASD, Sept5 in this cortical region does not seem to have any effect on social interaction.
Figure 4.
Effects of virally overexpressed Sept5 on active and passive social interaction in the dorsal hippocampus (A), basolateral amygdaloid complex (B), or somatosensory cortex (C). **and*, significant at 1 and 5% level, as determined by Newman-Keuls comparisons. This figure is reproduced from [109] with permission of the Oxford Press.
Our observations indicate that Sept5 is indeed a determinant of social interaction, but alleles in the genetic background may modulate phenotypic expression of 22q11.2-associated syndromic ASD. Consistent with our mouse phenotype, one child has recently been identified with homozygous deletion of Sept5 and adjacent GP1BB. This child exhibited deficits in motor development, social and emotional function as well as language and speech development [132]. Both parents were heterozygous with no apparent neuropsychiatric phenotypes. Moreover, the impacts of Sept5 expression in the mouse brain are largely consistent with human studies that underscore the critical roles played by limbic structures in ASD. Structural abnormalities in the hippocampus have been noted in both idiopathic ASD [127,128] and 22q11.2 hemizygous patients [129]. In individuals with 22q11.2 hemizygosity, amygdala activity is anomalous while performing tasks that require social perception [130]. A future challenge is to identify the precise network of structures through which Sept5 acts as a determinant for social cognition.
One interesting matter is that over-expression of Sept5 in the hippocampus and constitutive deletion of Sept5 increases and reduces social interaction, respectively. In an apparent contrast to this observation, clinical observations show that both duplication and hemizygosity induce similar behavioral phenotypes (e.g., social interaction deficits) in humans. However, while over-expression and hemizygosity of Tbx1 have been shown to induce similar cardiovascular phenotypes [131], it is not known if all individual 22q11.2 genes follow the same pattern in behavioral phenotypes. Thus, one possible explanation of our finding is that the dose level of some 22q11.2 genes linearly determines social interaction, but others do so in an inverted U gene-response curve. The phenotypic outcome of 22q11.2 hemizygosity and duplication might reflect the net effect of these additive or opposing phenotypes of multiple genes.
Conclusion
Systematic searches for 22q11.2 genes that contribute to behavioral phenotypes have identified potential significance of a ~200 kb segment. Data from our genetic mouse models suggest that Tbx1 and Sept5 within this ~200 kb region impact multiple or single symptomatic elements of 22q11.2-associated ASD. As Tbx1 is likely to be involved in postnatal neurogenesis [101] and Sept5 in synaptic contact [133–135] and neurotransmission [121,135,136], these neuronal events should be further explored as potential neuronal substrates for 22q11.2-associated ASD.
Acknowledgments
We thank the National Institute of Health (HD05311), NARSAD Independent Investigator Award, and the Maltz Foundation for their generous support. Permission has been granted from the Oxford Press for reproduction of our own published figures [101,108,109].
References
- 1.Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60:1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 2.Steffenburg S, Gillberg C, Hellgren L, Andersson L, Gillberg IC, et al. A twin study of autism in Denmark, Finland, Iceland, Norway and Sweden. J Child Psychol Psychiatry. 1989;30:405–416. doi: 10.1111/j.1469-7610.1989.tb00254.x. [DOI] [PubMed] [Google Scholar]
- 3.Ronald A, Happe F, Bolton P, Butcher LM, Price TS, et al. Genetic heterogeneity between the three components of the autism spectrum: a twin study. J Am Acad Child Adolesc Psychiatry. 2006;45:691–699. doi: 10.1097/01.chi.0000215325.13058.9d. [DOI] [PubMed] [Google Scholar]
- 4.Ronald A, Hoekstra RA. Autism spectrum disorders and autistic traits: a decade of new twin studies. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:255–274. doi: 10.1002/ajmg.b.31159. [DOI] [PubMed] [Google Scholar]
- 5.Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, et al. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med. 1995;25:63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- 6.Hallmayer J, Cleveland S, Torres A, Phillips J, Cohen B, et al. Genetic heritability and shared environmental factors among twin pairs with autism. Arch Gen Psychiatry. 2011;68:1095–1102. doi: 10.1001/archgenpsychiatry.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abrahams BS. Many Roads to the Autism Spectrum Disorders. Genetics. 2011:35–46. [Google Scholar]
- 8.Baker KD, Skuse DH. Adolescents and young adults with 22q11 deletion syndrome: psychopathology in an at-risk group. Br J Psychiatry. 2005;186:115–120. doi: 10.1192/bjp.186.2.115. [DOI] [PubMed] [Google Scholar]
- 9.Golding-Kushner KJ, Weller G, Shprintzen RJ. Velo-cardio-facial syndrome: language and psychological profiles. J Craniofac Genet Dev Biol. 1985;5:259–266. [PubMed] [Google Scholar]
- 10.Heineman-de Boer JA, Van Haelst MJ, Cordia-de Haan M, Beemer FA. Behavior problems and personality aspects of 40 children with velo-cardio-facial syndrome. Genet Couns. 1999;10:89–93. [PubMed] [Google Scholar]
- 11.Kiley-Brabeck K, Sobin C. Social skills and executive function deficits in children with the 22q11 Deletion Syndrome. Appl Neuropsychol. 2006;13:258–268. doi: 10.1207/s15324826an1304_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niklasson L, Rasmussen P, Oskarsdóttir S, Gillberg C. Chromosome 22q11 deletion syndrome (CATCH 22): neuropsychiatric and neuropsychological aspects. Dev Med Child Neurol. 2002;44:44–50. doi: 10.1017/s0012162201001645. [DOI] [PubMed] [Google Scholar]
- 13.Swillen A, Devriendt K, Legius E, Eyskens B, Dumoulin M, et al. Intelligence and psychosocial adjustment in velocardiofacial syndrome: a study of 37 children and adolescents with VCFS. J Med Genet. 1997;34:453–458. doi: 10.1136/jmg.34.6.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woodin M, Wang PP, Aleman D, McDonald-McGinn D, Zackai E, et al. Neuropsychological profile of children and adolescents with the 22q11.2 microdeletion. Genet Med. 2001;3:34–39. doi: 10.1097/00125817-200101000-00008. [DOI] [PubMed] [Google Scholar]
- 15.Shashi V, Veerapandiyan A, Schoch K, Kwapil T, Keshavan M, et al. Social skills and associated psychopathology in children with chromosome 22q11.2 deletion syndrome: implications for interventions. J Intellect Disabil Res. 2012;56:865–878. doi: 10.1111/j.1365-2788.2011.01477.x. [DOI] [PubMed] [Google Scholar]
- 16.Fine SE, Weissman A, Gerdes M, Pinto-Martin J, Zackai EH, et al. Autism spectrum disorders and symptoms in children with molecularly confirmed 22q11.2 deletion syndrome. J Autism Dev Disord. 2005;35:461–470. doi: 10.1007/s10803-005-5036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vorstman JA, Morcus ME, Duijff SN, Klaassen PW, Heineman-de Boer JA, et al. The 22q11.2 deletion in children: high rate of autistic disorders and early onset of psychotic symptoms. J Am Acad Child and Adolescent Psychiatry. 2006;45:1104–1113. doi: 10.1097/01.chi.0000228131.56956.c1. [DOI] [PubMed] [Google Scholar]
- 18.Antshel KM, Aneja A, Strunge L, Peebles J, Fremont WP, et al. Autistic spectrum disorders in velo-cardio facial syndrome (22q11.2 deletion) J Autism Dev Disord. 2007;37:1776–1786. doi: 10.1007/s10803-006-0308-6. [DOI] [PubMed] [Google Scholar]
- 19.Kates WR, Antshel KM, Fremont WP, Shprintzen RJ, Strunge LA, et al. Comparing phenotypes in patients with idiopathic autism to patients with velocardiofacial syndrome (22q11 DS) with and without autism. Am J Med Genet A. 2007;143A:2642–2650. doi: 10.1002/ajmg.a.32012. [DOI] [PubMed] [Google Scholar]
- 20.Niklasson L, Rasmussen P, Oskarsdottir S, Gillberg C. Autism, ADHD, mental retardation and behavior problems in 100 individuals with 22q11 deletion syndrome. Res Dev Disabil. 2009;30:763–773. doi: 10.1016/j.ridd.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 21.Esterberg ML, Ousley OY, Cubells JF, Walker EF. Prodromal and Autistic Symptoms in Schizotypal Personality Disorder and 22q11.2 Deletion Syndrome. J Abnorm Psychol. 2012 doi: 10.1037/a0028373. [DOI] [PubMed] [Google Scholar]
- 22.Lo-Castro A, Galasso C, Cerminara C, El-Malhany N, Benedetti S, et al. Association of syndromic mental retardation and autism with 22q11.2 duplication. Neuropediatrics. 2009;40:137–140. doi: 10.1055/s-0029-1237724. [DOI] [PubMed] [Google Scholar]
- 23.Mukaddes NM, Herguner S. Autistic disorder and 22q11.2 duplication. World J Biol Psychiatry. 2007;8:127–130. doi: 10.1080/15622970601026701. [DOI] [PubMed] [Google Scholar]
- 24.Ramelli GP, Silacci C, Ferrarini A, Cattaneo C, Visconti P, et al. Microduplication 22q11.2 in a child with autism spectrum disorder: clinical and genetic study. Dev Med Child Neurol. 2008;50:953–955. doi: 10.1111/j.1469-8749.2008.03048.x. [DOI] [PubMed] [Google Scholar]
- 25.Van Campenhout S, Devriendt K, Breckpot J, Frijns JP, Peeters H, et al. Microduplication 22q11.2: a description of the clinical, developmental and behavioral characteristics during childhood. Genet Couns. 2012;23:135–147. [PubMed] [Google Scholar]
- 26.Bucan M, Abrahams BS, Wang K, Glessner JT, Herman EI, et al. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet. 2009;5:e1000536. doi: 10.1371/journal.pgen.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai G, Edelmann L, Goldsmith JE, Cohen N, Nakamine A, et al. Multiplex ligation-dependent probe amplification for genetic spectrum disorders: efficient identification of known microduplications and identification of a novel microduplication in ASMT. BMC Med Genomics. 2008;1:50. doi: 10.1186/1755-8794-1-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christian SL, Brune CW, Sudi J, Kumar RA, Liu S, et al. Novel submicroscopic chromosomal abnormalities detected in autism spectrum disorder. Biol Psychiatry. 2008;63:1111–1117. doi: 10.1016/j.biopsych.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guilmatre A, Dubourg C, Mosca AL, Legallic S, Goldenberg A, et al. Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch Gen Psychiatry. 2009;66:947–956. doi: 10.1001/archgenpsychiatry.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Itsara A, Wu H, Smith JD, Nickerson DA, Romieu I, et al. De novo rates and selection of large copy number variation. Genome Res. 2010;20:1469–1481. doi: 10.1101/gr.107680.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bassett AS, Hodgkinson K, Chow EW, Correia S, Scutt LE, et al. 22q11 deletion syndrome in adults with schizophrenia. Am J Med Genet. 1998;81:328–337. [PMC free article] [PubMed] [Google Scholar]
- 36.Gothelf D, Frisch A, Munitz H, Rockah R, Laufer N, et al. Clinical characteristics of schizophrenia associated with velo-cardio-facial syndrome. Schizophr Res. 1999;35:105–112. doi: 10.1016/s0920-9964(98)00114-5. [DOI] [PubMed] [Google Scholar]
- 37.Moss EM, Batshaw ML, Solot CB, Gerdes M, McDonald-McGinn DM, et al. Psychoeducational profile of the 22q11.2 microdeletion: A complex pattern. J Pediatr. 1999;134:193–198. doi: 10.1016/s0022-3476(99)70415-4. [DOI] [PubMed] [Google Scholar]
- 38.Niklasson L, Rasmussen P, Oskarsdottir S, Gillberg C. Neuropsychiatric disorders in the 22q11 deletion syndrome. Genet Med. 2001;3:79–84. doi: 10.1097/00125817-200101000-00017. [DOI] [PubMed] [Google Scholar]
- 39.Swillen A, Devriendt K, Legius E, Prinzie P, Vogels A, et al. The behavioural phenotype in velo-cardio-facial syndrome (VCFS): from infancy to adolescence. Genet Couns. 1999;10:79–88. [PubMed] [Google Scholar]
- 40.Arnold PD, Siegel-Bartelt J, Cytrynbaum C, Teshima I, Schachar R. Velo-cardio-facial syndrome: Implications of microdeletion 22q11 for schizophrenia and mood disorders. Am J Med Genet. 2001;105:354–362. doi: 10.1002/ajmg.1359. [DOI] [PubMed] [Google Scholar]
- 41.Carlson C, Papolos D, Pandita RK, Faedda GL, Veit S, et al. Molecular analysis of velo-cardio-facial syndrome patients with psychiatric disorders. Am J Hum Genet. 1997;60:851–859. [PMC free article] [PubMed] [Google Scholar]
- 42.Feinstein C, Eliez S, Blasey C, Reiss AL. Psychiatric disorders and behavioral problems in children with velocardiofacial syndrome: usefulness as phenotypic indicators of schizophrenia risk. Biol Psychiatry. 2002;51:312–318. doi: 10.1016/s0006-3223(01)01231-8. [DOI] [PubMed] [Google Scholar]
- 43.Gothelf D, Presburger G, Zohar AH, Burg M, Nahmani A, et al. Obsessive-compulsive disorder in patients with velocardiofacial (22q11 deletion) syndrome. Am J Med Genet B Neuropsychiatr Genet. 2004;126:99–105. doi: 10.1002/ajmg.b.20124. [DOI] [PubMed] [Google Scholar]
- 44.Papolos DF, Faedda GL, Veit S, Goldberg R, Morrow B, et al. Bipolar spectrum disorders in patients diagnosed with velo-cardio-facial syndrome: does a hemizygous deletion of chromosome 22q11 result in bipolar affective disorder? Am J Psychiatry. 1996;153:1541–1547. doi: 10.1176/ajp.153.12.1541. [DOI] [PubMed] [Google Scholar]
- 45.Pulver AE, Nestadt G, Goldberg R, Shprintzen RJ, Lamacz M, et al. Psychotic illness in patients diagnosed with velo-cardio-facial syndrome and their relatives. J Nerv Ment Dis. 1994;182:476–478. doi: 10.1097/00005053-199408000-00010. [DOI] [PubMed] [Google Scholar]
- 46.Bassett AS, Chow EW. Schizophrenia and 22q11.2 deletion syndrome. Curr Psychiatry Rep. 2008;10:148–157. doi: 10.1007/s11920-008-0026-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shprintzen RJ, Goldberg R, Golding-Kushner KJ, Marion RW. Late-onset psychosis in the velo-cardio-facial syndrome. Am J Med Genet. 1992;42:141–142. doi: 10.1002/ajmg.1320420131. [DOI] [PubMed] [Google Scholar]
- 48.Murphy KC, Jones LA, Owen MJ. High rates of schizophrenia in adults with velo-cardio-facial syndrome. Arch Gen Psychiatry. 1999;56:940–945. doi: 10.1001/archpsyc.56.10.940. [DOI] [PubMed] [Google Scholar]
- 49.Karayiorgou M, Morris MA, Morrow B, Shprintzen RJ, Goldberg R, et al. Schizophrenia susceptibility associated with interstitial deletions of chromosome 22q11. Proc Natl Acad Sci U S A. 1995;92:7612–7616. doi: 10.1073/pnas.92.17.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bassett AS, Marshall CR, Lionel AC, Chow EW, Scherer SW. Copy number variations and risk for schizophrenia in 22q11.2 deletion syndrome. Hum Mol Genet. 2008;17:4045–4053. doi: 10.1093/hmg/ddn307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Debbane M, Glaser B, David MK, Feinstein C, Eliez S. Psychotic symptoms in children and adolescents with 22q11.2 deletion syndrome: Neuropsychological and behavioral implications. Schizophr Res. 2006;84:187–193. doi: 10.1016/j.schres.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 52.Raux G, Bumsel E, Hecketsweiler B, van Amelsvoort T, Zinkstok J, et al. Involvement of hyperprolinemia in cognitive and psychiatric features of the 22q11 deletion syndrome. Hum Mol Genet. 2007;16:83–91. doi: 10.1093/hmg/ddl443. [DOI] [PubMed] [Google Scholar]
- 53.Karayiorgou M, Simon TJ, Gogos JA. 22q11.2 microdeletions: linking DNA structural variation to brain dysfunction and schizophrenia. Nat Rev Neurosci. 2010;11:402–416. doi: 10.1038/nrn2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Green T, Gothelf D, Glaser B, Debbane M, Frisch A, et al. Psychiatric disorders and intellectual functioning throughout development in velocardiofacial (22q11.2 deletion) syndrome. J Am Acad Child Adolesc Psychiatry. 2009;48:1060–1068. doi: 10.1097/CHI.0b013e3181b76683. [DOI] [PubMed] [Google Scholar]
- 55.Eliez S. Autism in children with 22q11.2 deletion syndrome. J Am Acad Child Adolesc Psychiatry. 2007;46:433–434. doi: 10.1097/CHI.0b013e31802f5490. [DOI] [PubMed] [Google Scholar]
- 56.De Smedt B, Devriendt K, Fryns JP, Vogels A, Gewillig M, et al. Intellectual abilities in a large sample of children with Velo-Cardio-Facial Syndrome: an update. J Intellect Disabil Res. 2007;51:666–670. doi: 10.1111/j.1365-2788.2007.00955.x. [DOI] [PubMed] [Google Scholar]
- 57.Duijff SN, Klaassen PW, de Veye HF, Beemer FA, Sinnema G, et al. Cognitive development in children with 22q11.2 deletion syndrome. Br J Psychiatry. 2012;200:462–468. doi: 10.1192/bjp.bp.111.097139. [DOI] [PubMed] [Google Scholar]
- 58.Baird G, Charman T, Pickles A, Chandler S, Loucas T, et al. Regression, developmental trajectory and associated problems in disorders in the autism spectrum: the SNAP study. J Autism Dev Disord. 2008;38:1827–1836. doi: 10.1007/s10803-008-0571-9. [DOI] [PubMed] [Google Scholar]
- 59.Kjellmer L, Hedvall A, Fernell E, Gillberg C, Norrelgen F. Language and communication skills in preschool children with autism spectrum disorders: contribution of cognition, severity of autism symptoms, and adaptive functioning to the variability. Res Dev Disabil. 2012;33:172–180. doi: 10.1016/j.ridd.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 60.Smith V, Mirenda P, Zaidman-Zait A. Predictors of expressive vocabulary growth in children with autism. J Speech Lang Hear Res. 2007;50:149–160. doi: 10.1044/1092-4388(2007/013). [DOI] [PubMed] [Google Scholar]
- 61.Chevallier C, Kohls G, Troiani V, Brodkin ES, Schultz RT. The social motivation theory of autism. Trends Cogn Sci. 2012;16:231–239. doi: 10.1016/j.tics.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Senju A, Southgate V, White S, Frith U. Mindblind eyes: an absence of spontaneous theory of mind in Asperger syndrome. Science. 2009;325:883–885. doi: 10.1126/science.1176170. [DOI] [PubMed] [Google Scholar]
- 63.Scott-Van Zeeland AA, Dapretto M, Ghahremani DG, Poldrack RA, Bookheimer SY. Reward processing in autism. Autism Res. 2010;3:53–67. doi: 10.1002/aur.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Frith U. Mind blindness and the brain in autism. Neuron. 2001;32:969–979. doi: 10.1016/s0896-6273(01)00552-9. [DOI] [PubMed] [Google Scholar]
- 65.Baron-Cohen S, Jolliffe T, Mortimore C, Robertson M. Another advanced test of theory of mind: evidence from very high functioning adults with autism or asperger syndrome. J Child Psychol Psychiatry. 1997;38:813–822. doi: 10.1111/j.1469-7610.1997.tb01599.x. [DOI] [PubMed] [Google Scholar]
- 66.Fombonne E. The epidemiology of autism: a review. Psychol Med. 1999;29:769–786. doi: 10.1017/s0033291799008508. [DOI] [PubMed] [Google Scholar]
- 67.Fombonne E. Epidemiological surveys of autism and other pervasive developmental disorders: an update. J Autism Dev Disord. 2003;33:365–382. doi: 10.1023/a:1025054610557. [DOI] [PubMed] [Google Scholar]
- 68.Gillberg C, Ehlers S, Schaumann H, Jakobsson G, Dahlgren SO, et al. Autism under age 3 years: a clinical study of 28 cases referred for autistic symptoms in infancy. J Child Psychol Psychiatry. 1990;31:921–934. doi: 10.1111/j.1469-7610.1990.tb00834.x. [DOI] [PubMed] [Google Scholar]
- 69.Lord C, Volkmar F. Genetics of childhood disorders: XLII. Autism, part 1: Diagnosis and assessment in autistic spectrum disorders. J Am Acad Child Adolesc Psychiatry. 2002;41:1134–1136. doi: 10.1097/00004583-200209000-00015. [DOI] [PubMed] [Google Scholar]
- 70.Autism and Developmental Disabilities Monitoring Network Surveillance Year 2006 Principal Investigators. Prevalence of autism spectrum disorders - Autism and Developmental Disabilities Monitoring Network, United States, 2006. MMWR Surveill Summ. 2009;58:1–20. [PubMed] [Google Scholar]
- 71.Goldberg R, Motzkin B, Marion R, Scambler PJ, Shprintzen RJ. Velo-cardio-facial syndrome: a review of 120 patients. Am J Med Genet. 1993;45:313–319. doi: 10.1002/ajmg.1320450307. [DOI] [PubMed] [Google Scholar]
- 72.Swillen A, Vandeputte L, Cracco J, Maes B, Ghesquiere P, et al. Neuropsychological, learning and psychosocial profile of primary school aged children with the velo-cardio-facial syndrome (22q11 deletion): evidence for a nonverbal learning disability? Child Neuropsychol. 1999;5:230–241. doi: 10.1076/0929-7049(199912)05:04;1-R;FT230. [DOI] [PubMed] [Google Scholar]
- 73.Ogilvie CM, Moore J, Daker M, Palferman S, Docherty Z. Chromosome 22q11 deletions are not found in autistic patients identified using strict diagnostic criteria. IMGSAC. International Molecular Genetics Study of Autism Consortium. Am J Med Genet. 2000;96:15–17. [PubMed] [Google Scholar]
- 74.Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McDonald-McGinn DM, LaRossa D, Goldmuntz E, Sullivan K, Eicher P, et al. The 22q11.2 deletion: screening, diagnostic workup, and outcome of results; report on 181 patients. Genet Test. 1997;1:99–108. doi: 10.1089/gte.1997.1.99. [DOI] [PubMed] [Google Scholar]
- 76.Leana-Cox J, Pangkanon S, Eanet KR, Curtin MS, Wulfsberg EA. Familial DiGeorge/velocardiofacial syndrome with deletions of chromosome area 22q11.2: report of five families with a review of the literature. Am J Med Genet. 1996;65:309–316. doi: 10.1002/(SICI)1096-8628(19961111)65:4<309::AID-AJMG12>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 77.Ryan AK, Goodship JA, Wilson DI, Philip N, Levy A, et al. Spectrum of clinical features associated with interstitial chromosome 22q11 deletions: a European collaborative study. J Med Genet. 1997;34:798–804. doi: 10.1136/jmg.34.10.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Matsuoka R, Kimura M, Scambler PJ, Morrow BE, Imamura S, et al. Molecular and clinical study of 183 patients with conotruncal anomaly face syndrome. Hum Genet. 1998;103:70–80. doi: 10.1007/s004390050786. [DOI] [PubMed] [Google Scholar]
- 79.Coppinger J, McDonald-McGinn D, Zackai E, Shane K, Atkin JF, et al. Identification of familial and de novo microduplications of 22q11.21-q11.23 distal to the 22q11.21 microdeletion syndrome region. Hum Mol Genet. 2009;18:1377–1383. doi: 10.1093/hmg/ddp042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Courtens W, Schramme I, Laridon A. Microduplication 22q11.2: a benign polymorphism or a syndrome with a very large clinical variability and reduced penetrance?--Report of two families. Am J Med Genet A. 2008;146A:758–763. doi: 10.1002/ajmg.a.31910. [DOI] [PubMed] [Google Scholar]
- 81.Wincent J, Bruno DL, van Bon BW, Bremer A, Stewart H, et al. Sixteen New Cases Contributing to the Characterization of Patients with Distal 22q11.2 Microduplications. Mol Syndromol. 2010;1:246–254. doi: 10.1159/000327982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yu S, Cox K, Friend K, Smith S, Buchheim R, et al. Familial 22q11.2 duplication: a three-generation family with a 3-Mb duplication and a familial 1.5-Mb duplication. Clin Genet. 2008;73:160–164. doi: 10.1111/j.1399-0004.2007.00938.x. [DOI] [PubMed] [Google Scholar]
- 83.Unenge Hallerback M, Lugnegard T, Gillberg C. Is autism spectrum disorder common in schizophrenia? Psychiatry Res. 2012 doi: 10.1016/j.psychres.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 84.Mathieson I, Munafo MR, Flint J. Meta-analysis indicates that common variants at the DISC1 locus are not associated with schizophrenia. Mol Psychiatry. 2012;17:634–641. doi: 10.1038/mp.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hiroi N, Zhu H, Lee M, Funke B, Arai M, et al. A 200-kb region of human chromosome 22q11.2 confers antipsychotic-responsive behavioral abnormalities in mice. Proc Natl Acad Sci U S A. 2005;102:19132–19137. doi: 10.1073/pnas.0509635102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McDougle CJ, Stigler KA, Erickson CA, Posey DJ. A typical antipsychotics in children and adolescents with autistic and other pervasive developmental disorders. J Clin Psychiatry. 2008;69(Suppl 4):15–20. [PubMed] [Google Scholar]
- 87.Baker K, Baldeweg T, Sivagnanasundaram S, Scambler P, Skuse D. COMT Val108/158 Met modifies mismatch negativity and cognitive function in 22q11 deletion syndrome. Biol Psychiatry. 2005;58:23–31. doi: 10.1016/j.biopsych.2005.03.020. [DOI] [PubMed] [Google Scholar]
- 88.Campbell LE, Azuma R, Ambery F, Stevens A, Smith A, et al. Executive functions and memory abilities in children with 22q11.2 deletion syndrome. Aust N Z J Psychiatry. 2010;44:364–371. doi: 10.3109/00048670903489882. [DOI] [PubMed] [Google Scholar]
- 89.Goldenberg PC, Calkins ME, Richard J, McDonald-McGinn D, Zackai E, et al. Computerized neurocognitive profile in young people with 22q11.2 deletion syndrome compared to youths with schizophrenia and at-risk for psychosis. Am J Med Genet B Neuropsychiatr Genet. 2012;159B:87–93. doi: 10.1002/ajmg.b.32005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lajiness-O’Neill RR, Beaulieu I, Titus JB, Asamoah A, Bigler ED, et al. Memory and learning in children with 22q11.2 deletion syndrome: evidence for ventral and dorsal stream disruption? Child Neuropsychol. 2005;11:55–71. doi: 10.1080/09297040590911202. [DOI] [PubMed] [Google Scholar]
- 91.Lewandowski KE, Shashi V, Berry PM, Kwapil TR. Schizophrenic-like neurocognitive deficits in children and adolescents with 22q11 deletion syndrome. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:27–36. doi: 10.1002/ajmg.b.30379. [DOI] [PubMed] [Google Scholar]
- 92.O’Hearn K, Schroer E, Minshew N, Luna B. Lack of developmental improvement on a face memory task during adolescence in autism. Neuropsychologia. 2010;48:3955–3960. doi: 10.1016/j.neuropsychologia.2010.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Suzuki G, Harper KM, Hiramoto T, Funke B, Lee M, et al. Over-expression of a human chromosome 22q11.2 segment including TXNRD2, COMT, and ARVCF developmentally affects incentive learning and working memory in mice. Hum Mol Genet. 2009;18:3914–3925. doi: 10.1093/hmg/ddp334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stark KL, Burt RA, Gogos JA, Karayiorgou M. Analysis of prepulse inhibition in mouse lines overexpressing 22q11.2 orthologues. Int J Neuropsychopharmacol. 2009;12:983–989. doi: 10.1017/S1461145709000492. [DOI] [PubMed] [Google Scholar]
- 95.Papaleo F, Crawley JN, Song J, Lipska BK, Pickel J, et al. Genetic dissection of the role of catechol-O-methyltransferase in cognition and stress reactivity in mice. J Neurosci. 2008;28:8709–8723. doi: 10.1523/JNEUROSCI.2077-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sobin C, Kiley-Brabeck K, Karayiorgou M. Lower prepulse inhibition in children with the 22q11 deletion syndrome. Am J Psychiatry. 2005;162:1090–1099. doi: 10.1176/appi.ajp.162.6.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Paylor R, Glaser B, Mupo A, Ataliotis P, Spencer C, et al. Tbx1 haploinsufficiency is linked to behavioral disorders in mice and humans: implications for 22q11 deletion syndrome. Proc Natl Acad Sci USA. 2006;103:7729–7734. doi: 10.1073/pnas.0600206103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kimber WL, Hsieh P, Hirotsune S, Yuva-Paylor L, Sutherland HF, et al. Deletion of 150 kb in the minimal DiGeorge/velocardiofacial syndrome critical region in mouse. Hum Mol Genet. 1999;8:2229–2237. doi: 10.1093/hmg/8.12.2229. [DOI] [PubMed] [Google Scholar]
- 99.Long JM, LaPorte P, Merscher S, Funke B, Saint-Jore B, et al. Behavior of mice with mutations in the conserved region deleted in velocardiofacial/DiGeorge syndrome. Neurogenetics. 2006;7:247–257. doi: 10.1007/s10048-006-0054-0. [DOI] [PubMed] [Google Scholar]
- 100.Stark KL, Xu B, Bagchi A, Lai WS, Liu H, et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genet. 2008;40:751–760. doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- 101.Hiramoto T, Kang G, Suzuki G, Satoh Y, Kucherlapati R, et al. Tbx1: identification of a 22q11.2 gene as a risk factor for autism spectrum disorder in a mouse model. Hum Mol Genet. 2011;20:4775–4785. doi: 10.1093/hmg/ddr404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Meechan DW, Maynard TM, Wu Y, Gopalakrishna D, Lieberman JA, et al. Gene dosage in the developing and adult brain in a mouse model of 22q11 deletion syndrome. Mol Cell Neurosci. 2006;33:412–428. doi: 10.1016/j.mcn.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 103.Geyer MA. The family of sensorimotor gating disorders: comorbidities or diagnostic overlaps? Neurotox Res. 2006;10:211–220. doi: 10.1007/BF03033358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ornitz EM, Lane SJ, Sugiyama T, deTraversay J. Startle modulation studies in autism. J Autism Dev Disord. 1993;23:619–637. doi: 10.1007/BF01046105. [DOI] [PubMed] [Google Scholar]
- 105.Perry W, Minassian A, Lopez B, Maron L, Lincoln A. Sensorimotor gating deficits in adults with autism. Biol Psychiatry. 2007;61:482–486. doi: 10.1016/j.biopsych.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 106.Yuhas J, Cordeiro L, Tassone F, Ballinger E, Schneider A, et al. Brief Report: Sensorimotor Gating in Idiopathic Autism and Autism Associated with Fragile X Syndrome. J Autism Dev Disord. 2011;41:248–53. doi: 10.1007/s10803-010-1040-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.McAlonan GM, Daly E, Kumari V, Critchley HD, van Amelsvoort T, et al. Brain anatomy and sensorimotor gating in Asperger’s syndrome. Brain. 2002;125:1594–1606. doi: 10.1093/brain/awf150. [DOI] [PubMed] [Google Scholar]
- 108.Suzuki G, Harper KM, Hiramoto T, Sawamura T, Lee M, et al. Sept5 deficiency exerts pleiotropic influence on affective behaviors and cognitive functions in mice. Hum Mol Genet. 2009;18:1652–1660. doi: 10.1093/hmg/ddp086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Harper KM, Hiramoto T, Tanigaki K, Kang G, Suzuki G, et al. Alterations of social interaction through genetic and environmental manipulation of the 22q11.2 gene Sept5 in the mouse brain. Hum Mol Genet. 2012;121:3489–3499. doi: 10.1093/hmg/dds180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Harper KM. Doctoral thesis. Albert Einstein College of Medicine; 2012. Sept5: alteration of social behavior through genetic and environmental manipulation of a 22q11.2 gene in mice. [Google Scholar]
- 111.Crawley JN. Mouse behavioral assays relevant to the symptoms of autism. Brain Pathol. 2007;17:448–459. doi: 10.1111/j.1750-3639.2007.00096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Solot CB, Knightly C, Handler SD, Gerdes M, McDonald-McGinn DM, et al. Communication disorders in the 22Q11.2 microdeletion syndrome. J Commun Disord. 2000;33:187–203. doi: 10.1016/s0021-9924(00)00018-6. [DOI] [PubMed] [Google Scholar]
- 113.Scattoni ML, Crawley J, Ricceri L. Ultrasonic vocalizations: a tool for behavioural phenotyping of mouse models of neurodevelopmental disorders. Neurosci Biobehav Rev. 2009;33:508–515. doi: 10.1016/j.neubiorev.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lalonde R. The neurobiological basis of spontaneous alternation. Neurosci Biobehav Rev. 2002;26:91–104. doi: 10.1016/s0149-7634(01)00041-0. [DOI] [PubMed] [Google Scholar]
- 115.Kana RK, Keller TA, Minshew NJ, Just MA. Inhibitory control in high-functioning autism: decreased activation and underconnectivity in inhibition networks. Biol Psychiatry. 2007;62:198–206. doi: 10.1016/j.biopsych.2006.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.O’Hearn K, Asato M, Ordaz S, Luna B. Neurodevelopment and executive function in autism. Dev Psychopathol. 2008;20:1103–1132. doi: 10.1017/S0954579408000527. [DOI] [PubMed] [Google Scholar]
- 117.Caltagarone J, Rhodes J, Honer WG, Bowser R. Localization of a novel septin protein, hCDCrel-1, in neurons of human brain. Neuroreport. 1998;9:2907–2912. doi: 10.1097/00001756-199808240-00042. [DOI] [PubMed] [Google Scholar]
- 118.Kinoshita A, Noda M, Kinoshita M. Differential localization of septins in the mouse brain. J Comp Neurol. 2000;428:223–239. doi: 10.1002/1096-9861(20001211)428:2<223::aid-cne3>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 119.Beites CL, Xie H, Bowser R, Trimble WS. The septin CDCrel-1 binds syntaxin and inhibits exocytosis. Nat Neurosci. 1999;2:434–439. doi: 10.1038/8100. [DOI] [PubMed] [Google Scholar]
- 120.Yang YM, Fedchyshyn MJ, Grande G, Aitoubah J, Tsang CW, et al. Septins regulate developmental switching from microdomain to nanodomain coupling of Ca(2+) influx to neurotransmitter release at central synapse. Neuron. 2010;67:100–115. doi: 10.1016/j.neuron.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dong Z, Ferger B, Paterna JC, Vogel D, Furler S, et al. Dopamine-dependent neurodegeneration in rats induced by viral vector-mediated overexpression of the parkin target protein, CDCrel-1. Proc Natl Acad Sci USA. 2003;100:12438–12443. doi: 10.1073/pnas.2132992100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tada T, Simonetta A, Batterton M, Kinoshita M, Edbauer D, et al. Role of Septin cytoskeleton in spine morphogenesis and dendrite development in neurons. Curr Biol. 2007;17:1752–1758. doi: 10.1016/j.cub.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tsang CW, Estey MP, DiCiccio JE, Xie H, Patterson D, et al. Characterization of presynaptic septin complexes in mammalian hippocampal neurons. Biol Chem. 2011;392:739–749. doi: 10.1515/BC.2011.077. [DOI] [PubMed] [Google Scholar]
- 124.Flaherty L, Bolivar V. Congenic and consomic strains. 2. 2007. pp. 115–127. [Google Scholar]
- 125.Crusio WE. Flanking gene and genetic background problems in genetically manipulated mice. Biol Psychiatry. 2004;56:381–385. doi: 10.1016/j.biopsych.2003.12.026. [DOI] [PubMed] [Google Scholar]
- 126.Wolfer DP, Crusio WE, Lipp HP. Knockout mice: simple solutions to the problems of genetic background and flanking genes. Trends Neurosci. 2002;25:336–340. doi: 10.1016/s0166-2236(02)02192-6. [DOI] [PubMed] [Google Scholar]
- 127.Rojas DC, Smith JA, Benkers TL, Camou SL, Reite ML, et al. Hippocampus and amygdala volumes in parents of children with autistic disorder. Am J Psychiatry. 2004;161:2038–2044. doi: 10.1176/appi.ajp.161.11.2038. [DOI] [PubMed] [Google Scholar]
- 128.Rojas DC, Peterson E, Winterrowd E, Reite ML, Rogers SJ, et al. Regional gray matter volumetric changes in autism associated with social and repetitive behavior symptoms. BMC Psychiatry. 2006;6:56. doi: 10.1186/1471-244X-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tan GM, Arnone D, McIntosh AM, Ebmeier KP. Meta-analysis of magnetic resonance imaging studies in chromosome 22q11.2 deletion syndrome (velocardiofacial syndrome) Schizophr Res. 2009;115:173–181. doi: 10.1016/j.schres.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 130.Andersson F, Glaser B, Spiridon M, Debbané M, Vuilleumier P, et al. Impaired activation of face processing networks revealed by functional magnetic resonance imaging in 22q11.2 deletion syndrome. Biol Psychiatry. 2008;63:49–57. doi: 10.1016/j.biopsych.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 131.Liao J, Kochilas L, Nowotschin S, Arnold JS, Aggarwal VS, et al. Full spectrum of malformations in velo-cardio-facial syndrome/DiGeorge syndrome mouse models by altering Tbx1 dosage. Hum Mol Genet. 2004;13:1577–1585. doi: 10.1093/hmg/ddh176. [DOI] [PubMed] [Google Scholar]
- 132.Bartsch I, Sandrock K, Lanza F, Nurden P, Hainmann I, et al. Deletion of human GP1BB and SEPT5 is associated with Bernard-Soulier syndrome, platelet secretion defect, polymicrogyria, and developmental delay. Thromb Haemost. 2011;106:475–483. doi: 10.1160/TH11-05-0305. [DOI] [PubMed] [Google Scholar]
- 133.Tada T, Simonetta A, Batterton M, Kinoshita M, Edbauer D, et al. Role of Septin cytoskeleton in spine morphogenesis and dendrite development in neurons. Curr Biol. 2007;17:1752–1758. doi: 10.1016/j.cub.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tsang CW, Estey MP, DiCiccio JE, Xie H, Patterson D, et al. Characterization of presynaptic septin complexes in mammalian hippocampal neurons. Biol Chem. 2011;392:739–749. doi: 10.1515/BC.2011.077. [DOI] [PubMed] [Google Scholar]
- 135.Beites CL, Xie H, Bowser R, Trimble WS. The septin CDCrel-1 binds syntaxin and inhibits exocytosis. Nat Neurosci. 1999;2:434–439. doi: 10.1038/8100. [DOI] [PubMed] [Google Scholar]
- 136.Yang YM, Fedchyshyn MJ, Grande G, Aitoubah J, Tsang CW, et al. Septins regulate developmental switching from microdomain to nanodomain coupling of Ca(2+) influx to neurotransmitter release at a central synapse. Neuron. 2010;67:100–115. doi: 10.1016/j.neuron.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Scattoni ML, Gandhy SU, Ricceri L, Crawley JN. Unusual repertoire of vocalizations in the BTBR T+tf/J mouse model of autism. PLoS One. 2008;3:e3067. doi: 10.1371/journal.pone.0003067. [DOI] [PMC free article] [PubMed] [Google Scholar]