Abstract
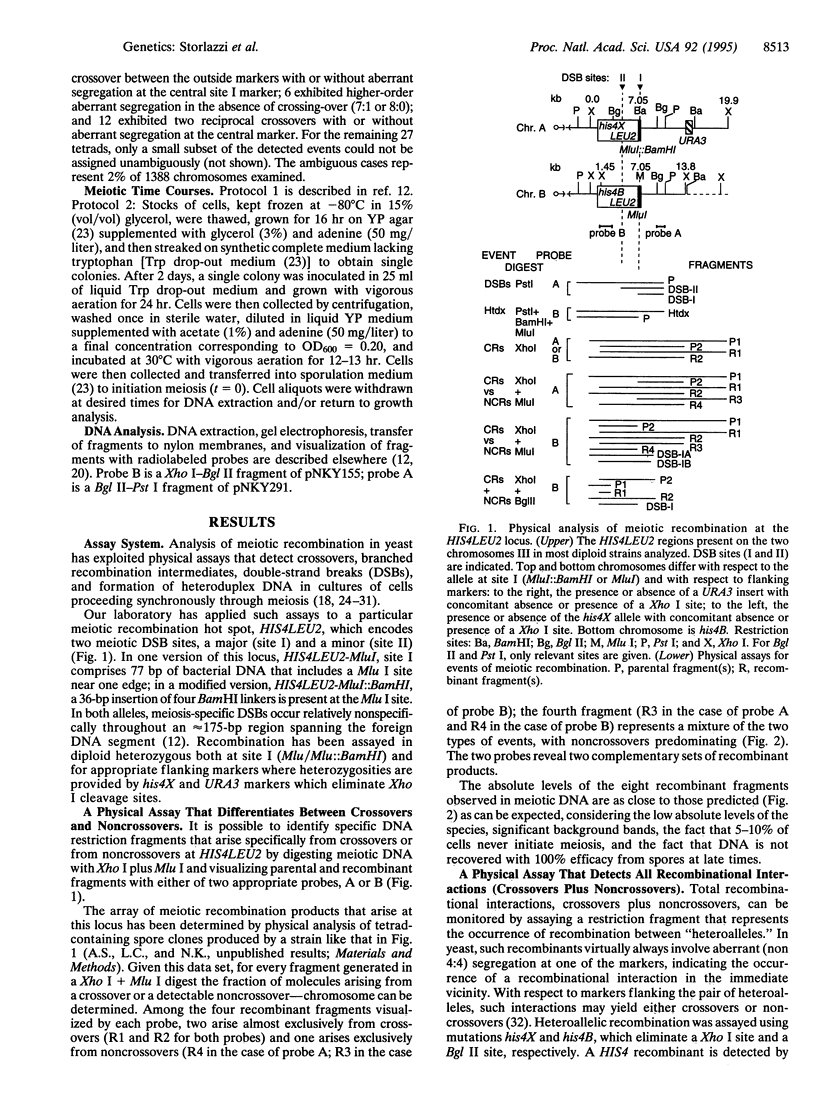
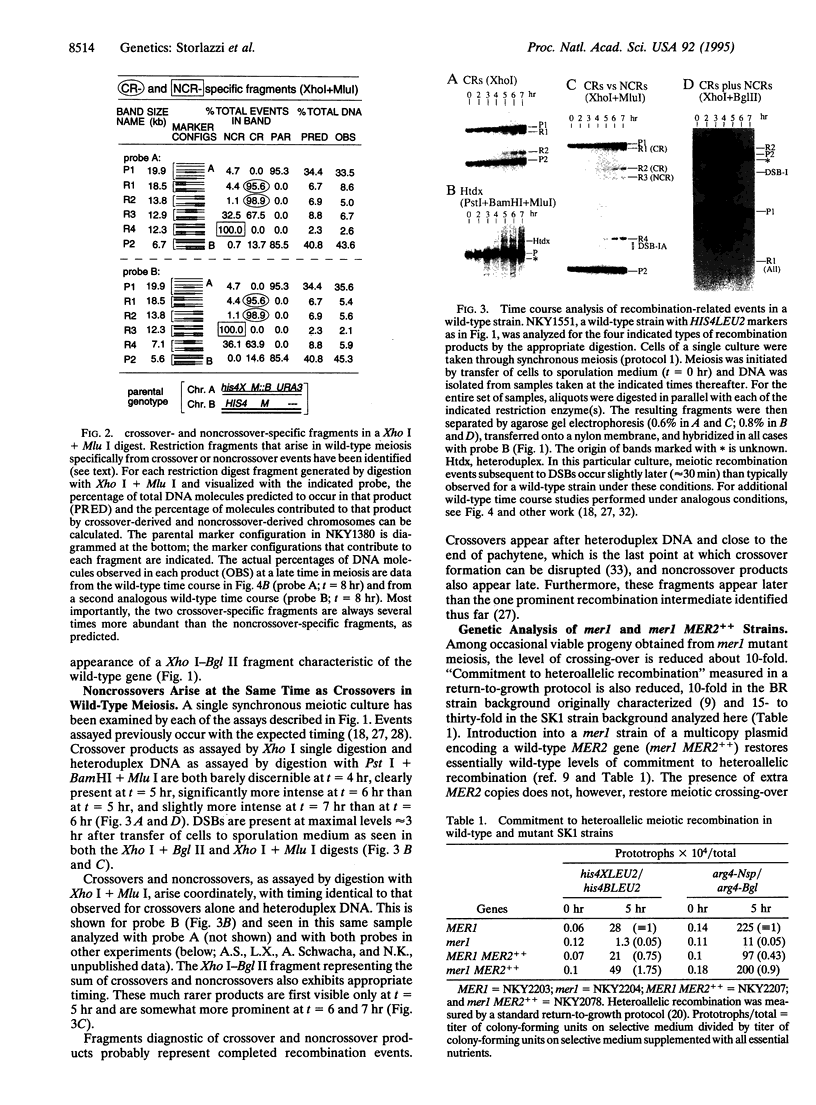
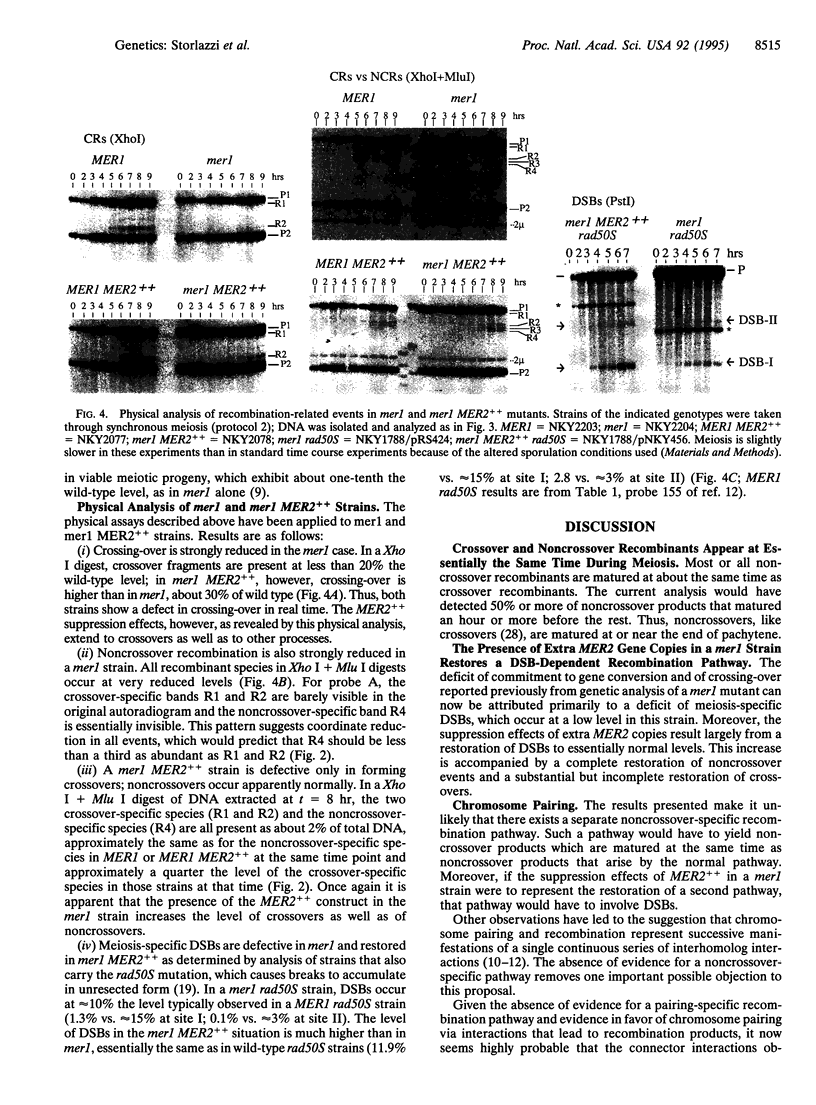
During meiosis, crossovers occur at a high level, but the level of noncrossover recombinants is even higher. The biological rationale for the existence of the latter events is not known. It has been suggested that a noncrossover-specific pathway exists specifically to mediate chromosome pairing. Using a physical assay that monitors both crossovers and noncrossovers in cultures of yeast undergoing synchronous meiosis, we find that both types of products appear at essentially the same time, after chromosomes are fully synapsed at pachytene. We have also analyzed a situation in which commitment to meiotic recombination and formation of the synaptonemal complex are coordinately suppressed (mer1 versus mer1 MER2++). We find that suppression is due primarily to restoration of meiosis-specific double-strand breaks, a characteristic of the major meiotic recombination pathway. Taken together, the observations presented suggest that there probably is no noncrossover-specific pathway and that restoration of intermediate events in a single pairing/recombination pathway promotes synaptonemal complex formation. The biological significant of noncrossover recombination remains to be determined, however.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alani E., Cao L., Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987 Aug;116(4):541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alani E., Padmore R., Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990 May 4;61(3):419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- Bell L. R., Byers B. Homologous association of chromosomal DNA during yeast meiosis. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):829–840. doi: 10.1101/sqb.1983.047.01.095. [DOI] [PubMed] [Google Scholar]
- Bishop D. K., Park D., Xu L., Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992 May 1;69(3):439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Borts R. H., Lichten M., Haber J. E. Analysis of meiosis-defective mutations in yeast by physical monitoring of recombination. Genetics. 1986 Jul;113(3):551–567. doi: 10.1093/genetics/113.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borts R. H., Lichten M., Hearn M., Davidow L. S., Haber J. E. Physical monitoring of meiotic recombination in Saccharomyces cerevisiae. Cold Spring Harb Symp Quant Biol. 1984;49:67–76. doi: 10.1101/sqb.1984.049.01.010. [DOI] [PubMed] [Google Scholar]
- Cao L., Alani E., Kleckner N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell. 1990 Jun 15;61(6):1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- Carpenter A. T. Chiasma function. Cell. 1994 Jul 1;77(7):957–962. doi: 10.1016/0092-8674(94)90434-0. [DOI] [PubMed] [Google Scholar]
- Carpenter A. T. Gene conversion, recombination nodules, and the initiation of meiotic synapsis. Bioessays. 1987 May;6(5):232–236. doi: 10.1002/bies.950060510. [DOI] [PubMed] [Google Scholar]
- Egel R. Synaptonemal complex and crossing-over: structural support or interference? Heredity (Edinb) 1978 Oct;41(2):233–237. doi: 10.1038/hdy.1978.92. [DOI] [PubMed] [Google Scholar]
- Engebrecht J., Hirsch J., Roeder G. S. Meiotic gene conversion and crossing over: their relationship to each other and to chromosome synapsis and segregation. Cell. 1990 Sep 7;62(5):927–937. doi: 10.1016/0092-8674(90)90267-i. [DOI] [PubMed] [Google Scholar]
- Engebrecht J., Roeder G. S. MER1, a yeast gene required for chromosome pairing and genetic recombination, is induced in meiosis. Mol Cell Biol. 1990 May;10(5):2379–2389. doi: 10.1128/mcb.10.5.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engebrecht J., Roeder G. S. Yeast mer1 mutants display reduced levels of meiotic recombination. Genetics. 1989 Feb;121(2):237–247. doi: 10.1093/genetics/121.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink G. R., Petes T. D. Gene conversion in the absence of reciprocal recombination. 1984 Aug 30-Sep 5Nature. 310(5980):728–729. doi: 10.1038/310728a0. [DOI] [PubMed] [Google Scholar]
- Foss E., Lande R., Stahl F. W., Steinberg C. M. Chiasma interference as a function of genetic distance. Genetics. 1993 Mar;133(3):681–691. doi: 10.1093/genetics/133.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyon C., Lichten M. Timing of molecular events in meiosis in Saccharomyces cerevisiae: stable heteroduplex DNA is formed late in meiotic prophase. Mol Cell Biol. 1993 Jan;13(1):373–382. doi: 10.1128/mcb.13.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson S. A. The time and place of meiotic crossing-over. Annu Rev Genet. 1970;4:295–324. doi: 10.1146/annurev.ge.04.120170.001455. [DOI] [PubMed] [Google Scholar]
- Kane S. M., Roth R. Carbohydrate metabolism during ascospore development in yeast. J Bacteriol. 1974 Apr;118(1):8–14. doi: 10.1128/jb.118.1.8-14.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J. S., Mortimer R. K. A polymerization model of chiasma interference and corresponding computer simulation. Genetics. 1990 Dec;126(4):1127–1138. doi: 10.1093/genetics/126.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N., Weiner B. M. Potential advantages of unstable interactions for pairing of chromosomes in meiotic, somatic, and premeiotic cells. Cold Spring Harb Symp Quant Biol. 1993;58:553–565. doi: 10.1101/sqb.1993.058.01.062. [DOI] [PubMed] [Google Scholar]
- Loidl J., Nairz K., Klein F. Meiotic chromosome synapsis in a haploid yeast. Chromosoma. 1991 May;100(4):221–228. doi: 10.1007/BF00344155. [DOI] [PubMed] [Google Scholar]
- Nag D. K., Petes T. D. Physical detection of heteroduplexes during meiotic recombination in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1993 Apr;13(4):2324–2331. doi: 10.1128/mcb.13.4.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmore R., Cao L., Kleckner N. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell. 1991 Sep 20;66(6):1239–1256. doi: 10.1016/0092-8674(91)90046-2. [DOI] [PubMed] [Google Scholar]
- Powers P. A., Smithies O. Short gene conversions in the human fetal globin gene region: a by-product of chromosome pairing during meiosis? Genetics. 1986 Feb;112(2):343–358. doi: 10.1093/genetics/112.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha A., Kleckner N. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell. 1994 Jan 14;76(1):51–63. doi: 10.1016/0092-8674(94)90172-4. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Michaud W. A., Wootton J. C., Boguski M. S., Connelly C., Hieter P. TPR proteins as essential components of the yeast cell cycle. Cold Spring Harb Symp Quant Biol. 1991;56:663–673. doi: 10.1101/sqb.1991.056.01.075. [DOI] [PubMed] [Google Scholar]
- Smithies O., Powers P. A. Gene conversions and their relation to homologous chromosome pairing. Philos Trans R Soc Lond B Biol Sci. 1986 Jan 29;312(1154):291–302. doi: 10.1098/rstb.1986.0008. [DOI] [PubMed] [Google Scholar]
- Sun H., Treco D., Schultes N. P., Szostak J. W. Double-strand breaks at an initiation site for meiotic gene conversion. Nature. 1989 Mar 2;338(6210):87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- Sym M., Engebrecht J. A., Roeder G. S. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 1993 Feb 12;72(3):365–378. doi: 10.1016/0092-8674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- Weiner B. M., Kleckner N. Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell. 1994 Jul 1;77(7):977–991. doi: 10.1016/0092-8674(94)90438-3. [DOI] [PubMed] [Google Scholar]
- Zickler D., Moreau P. J., Huynh A. D., Slezec A. M. Correlation between pairing initiation sites, recombination nodules and meiotic recombination in Sordaria macrospora. Genetics. 1992 Sep;132(1):135–148. doi: 10.1093/genetics/132.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]




