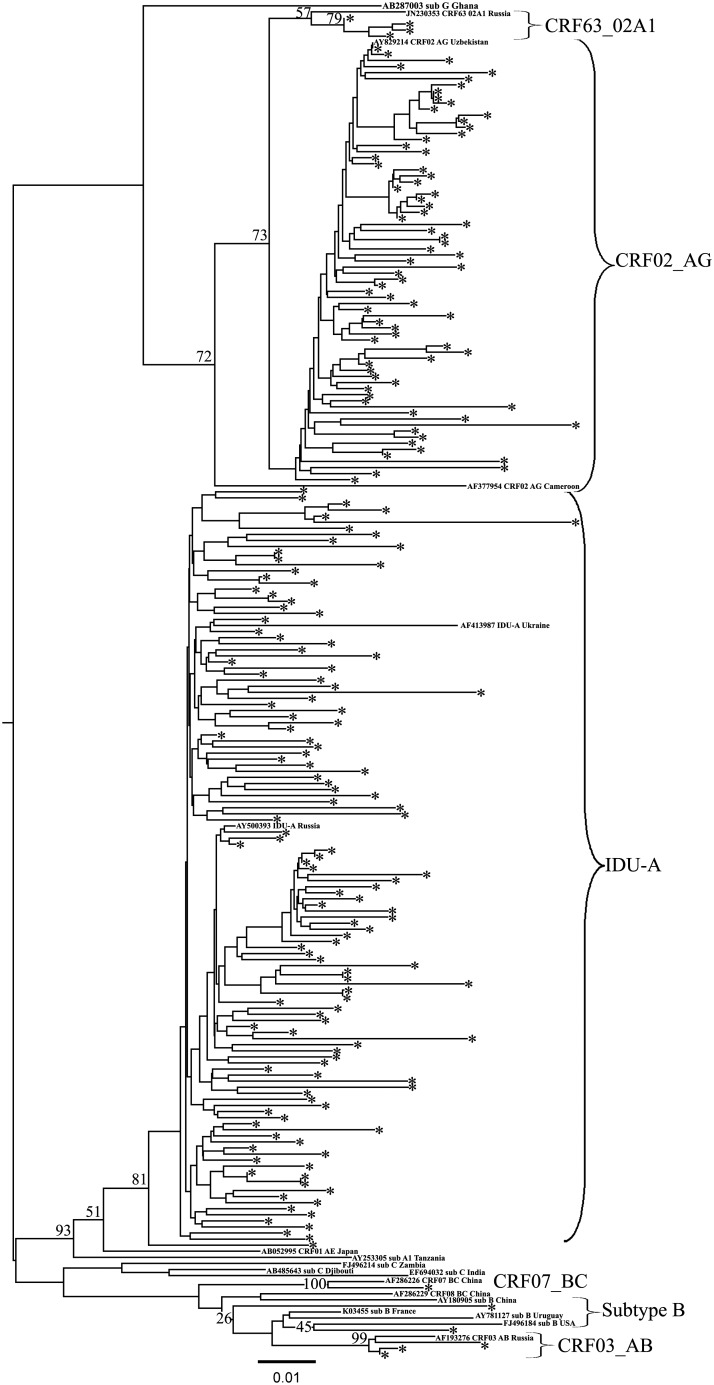
FIG. 2.
Phylogenetic analysis of HIV-1 pol sequences (joined Pro-RT region) from 2009–2013 study individuals. The phylogenetic tree was constructed using the PhyML tool by the maximum likelihood method of phylogeny, the bootstrap method of branch support (100 replicates), and the GTR + gamma nucleotide substitution model. The optimization of the phylogenetic tree was carried out by FigTree v.1.3.1. Sequences obtained in this study are labeled as snowflakes. Bootstrap values as percentages are indicated at the nodes.