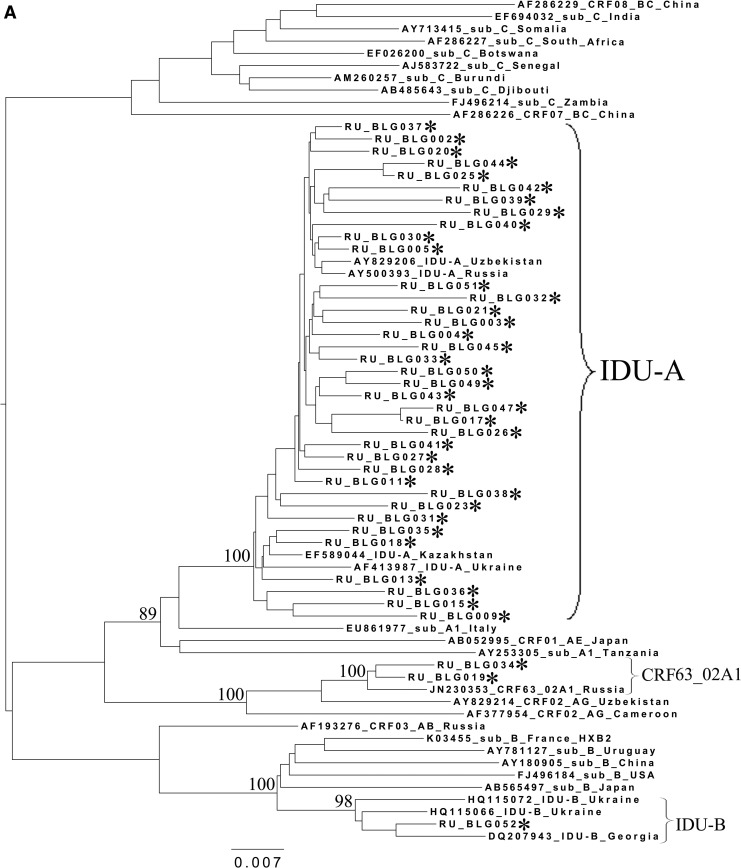
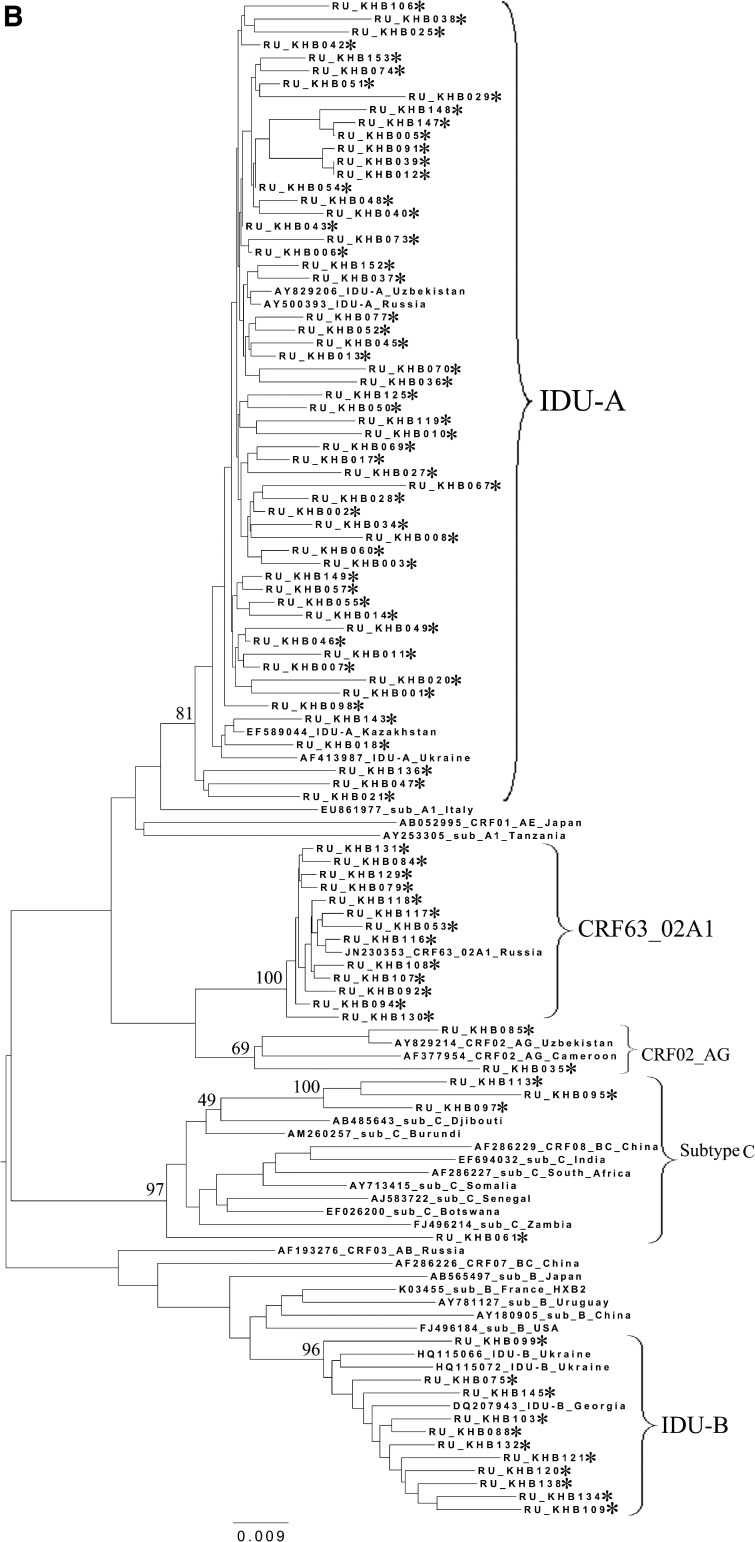
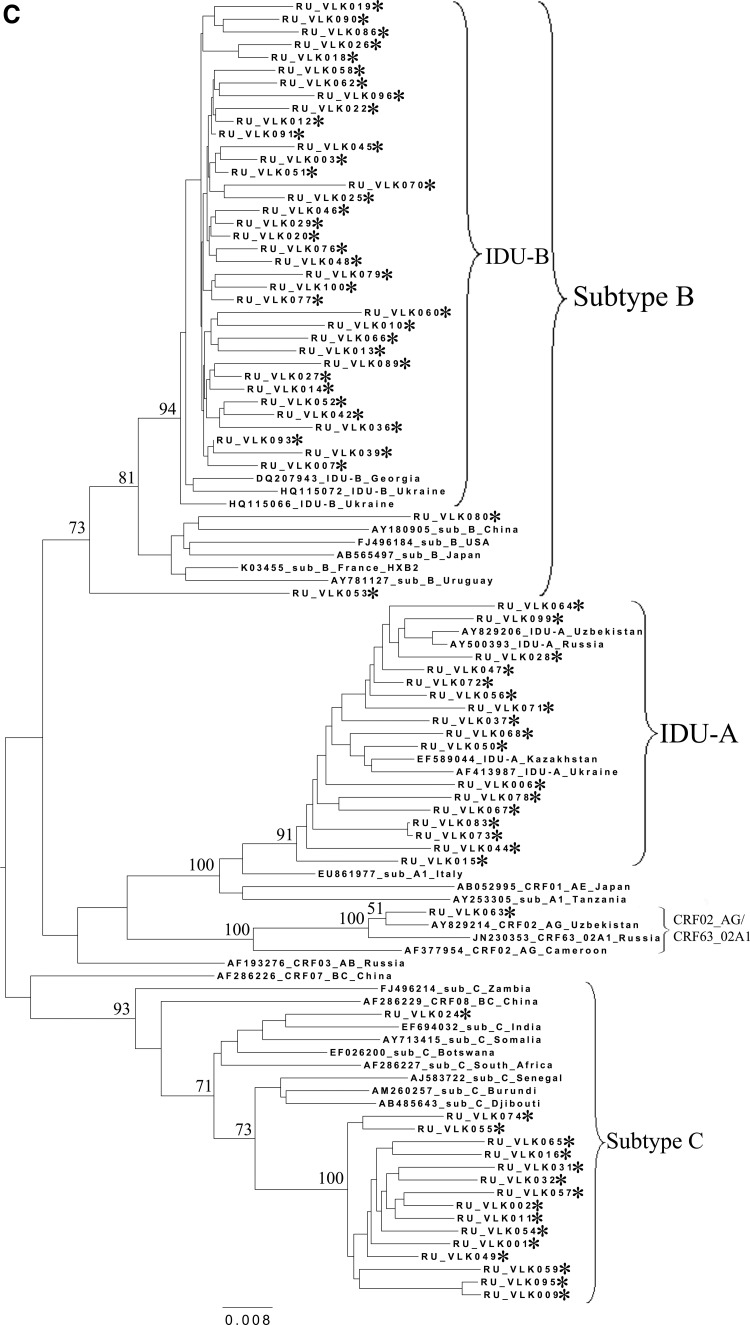
FIG. 2.
Phylogenetic analysis of HIV-1 pol sequences coding the Pro and reverse transcriptase (RT) regions from samples collected in Blagoveshchensk (2253–3554 bp) (A), Khabarovsk (2253–3554 bp) (B), and Vladivostok (2253–3167 bp) (C). A phylogenetic tree was constructed using the PhyML tool by the maximum likelihood method of phylogeny, the bootstrap method of branch support (100 replicates), and the GTR+gamma nucleotide substitution model. Optimization of the phylogenetic tree was carried out by FigTree v.1.3.1. Nucleotide positions are shown with respect to the HXB2 genome (GenBank number K03455). The sequences obtained in this study are marked with snowflakes. Reference isolates are represented by their subtypes and sampling countries. Node support values as percentages are indicated at the nodes.