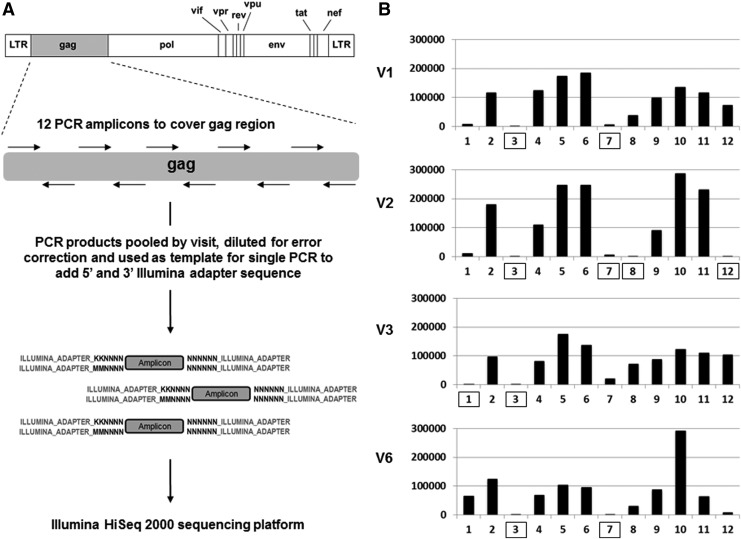
FIG. 2.
Deep sequencing. (A) Schematic of sample preparation for the deep sequencing approach. HIV-1 gag cDNA was used as a template for the first step PCR using 12 primer pairs containing overhangs with unique nucleotide tag and partial sequence corresponding to the Illumina adapter. PCR products were pooled by visit, diluted for error correction, and used in a single PCR reaction to add the remaining Illumina adapter sequence before deep sequencing. (B) Sequence coverage of each amplicon covering the HIV-1 gag region per visit. Box denotes amplicons where coverage was <5,000 error-corrected reads and not used in further analysis.