Abstract
Members of the Sox (Sry-related high mobility group box) family of transcription factors play a variety of roles during development of both vertebrates and invertebrates. A marked expansion in gene number occurred during emergence of vertebrates, apparently via gene duplication events that are thought to have facilitated new functions. By screening a macroarrayed library as well as the lamprey genome, we have isolated genes of the Sox B, D, E and F subfamilies in the basal jawless vertebrate, lamprey. The expression patterns of all identified Sox genes were examined from gastrulation through early organogenesis (embryonic day 4–14), with particular emphasis on the neural crest, a vertebrate innovation. Coupled with phylogenetic analysis of these Sox genes, the results provide insight into gene duplication and divergence in paralog deployment occurring during early vertebrate evolution.
Keywords: Sox Genes, lamprey, cyclostomes, neural crest
Introduction
Many transcription factors of the Sox gene family are critical for a number of developmental processes, most notably sex determination, neural crest development and neurogenesis (Laudet et al., 1993; Hong et al., 2005; Betancur et al., 2010). This family is comprised of more than 30 genes that have been classified into eight paralogy groups [SoxA-SoxH; (Schepers et al., 2002)]. Sox genes are characterized by the presence of a single High Mobility Group box (HMG box), a 79 amino acid DNA binding domain that has affinity to the WWCAAW motif (Laudet et al., 1993). These factors are generally expressed in a dynamic, tissue specific manner, and often interact with other transcription factors (Prior et al., 1996).
Members of the Sox gene family are found across the animal kingdom, and it has been suggested that the major Sox groups (i.e. B, C, D and E) were already present in the common bilaterian ancestor, whereas group A genes are specific to mammals (Jager et al., 2006). Expansion in the number of vertebrate Sox genes is thought to be due to major gene duplication events, initially occurring during early stages of metazoan evolution and later during the transition between non-vertebrate chordates and vertebrates (Prior et al., 1996; Dehal et al., 2005). Gnathostomes (jawed vertebrates) have undergone two rounds of whole genome duplications (Escriva et al., 2002), whereas estimates for the number of rounds of duplication in agnathans like lamprey range from one to two. It is not yet clear whether supernumerary copies of lamprey genes arose via whole genome-wide duplication, or via independent duplication events (Tomsa et al., 1999; McCauley et al., 2006; Neidert et al., 2001; Zhong et al., 2011).
Classification of Sox genes was first done by Wright et al., 1993, using partial sequences from mouse SOX genes. This study defined the six paralogous groups (A-F) which are the basis of the current classification (Wright et al., 1993). Four more groups were subsequently added to include recently identified paralogs (Bowles et al., 2000). Nevertheless, members of the same groups do not always have similar roles or expression patterns, indicating that recent paralogs can adopt new functions with relative ease (Bowles et al., 2000).
In the neural crest, a vertebrate innovation that contributes to the peripheral nervous system and craniofacial skeleton, the function of Sox genes has been studied at many stages. For example, gnathostome SoxE family members (Sox8, Sox9, and Sox10) are expressed in premigratory neural crest progenitors, migrating neural crest cells, as well as at later stages, in numerous neural crest derivatives (Sauka-Spengler et al., 2008), similar to their lamprey paralogs (McCaulsey et al., 2006) SoxD family members (Sox5 and Sox6) are found in cranial ganglia (Morales et al., 2007) and cartilage elements of neural crest origin. Additionally, gnathostome SoxB family members have been shown to play an essential role in differentiation of late neural crest derivatives (Wakamatsu et al., 2004).
Given the important function of Sox genes in gnathostomes, we sought to identify and characterize novel lamprey genes of the SoxD and SoxF subfamilies, as well as additional members of the SoxB and SoxE groups. As one of the basal-most extant vertebrates, analysis of the deployment of paralogous genes in lamprey offers the opportunity to examine events in early emergence of vertebrate specific features. Comparative amino acid analysis between lamprey and other vertebrate Sox genes provides insight into the evolutionary history of early vertebrates, as well as duplication events occurring early in the vertebrate lineage.
Materials and Methods
Heterospecific screening of an arrayed lamprey embryonic cDNA library
A high quality directional full-length arrayed cDNA library (Sauka-Spengler et al., 2007) from embryonic day 2–12 lamprey embryos (an average efficiency of ~0.9 × 108 transformants/μg of cDNA) was used for low-stringency screening. Nine individual nitrocellulose filters were screened using Sox heterospecific probes, yielding 7 different Sox genes (SoxB, E, and F family members), whose identity was confirmed by sequencing on both strands, BLAST searching and phylogenetic analysis (see below).
RNA-ligated mediated 5′ Rapid Amplification of cDNA ends (RLM-5′ RACE)
A SoxD homologue was identified by bioinformatic survey of the lamprey genomic sequences and cloned using RACE. RACE was also used to obtain full-length sequences of the Sox genes, where cDNA clones were incomplete. Total RNA was extracted from 6, 8, 10, and 14 day old embryos from Ambion:RNAquous kit. RLM-5′ RACE was conducted on the total mRNA in accordance with Invitrogen: GeneRacer Kit. Total RNA was dephosphorylated through Calf Intestinal Phosphatase (CIP) treatment, decapped via Tobacco Acid Pyrophosphatase (TAP), ligated with the GeneRacer RNA oligo, and finally reverse transcribed using random hexamer priming to form the cDNA template. Gene specific primers were:
GeneRacer 5′ Primer CGACTGGAGCACGAGGACACTGA
SoxB1-B: 5′ CGACTGGAGCACGAGGACACTGA 3′
SoxD: 5′ CGCCTCTCGTCCTTTGCCCAGAC3′
Touch Down PCR
The touchdown PCR procedure was based off of the Invitrogen GeneRacer Kit. Samples were prepared in 5% DMSO, using TAQ polymerase with Roche Expand Long Template PCR Buffer 1(10x concentrated, 17.5 mM MgCl2). A hot start at 94°C was conducted followed by 5 cycles of 94°C for 30 seconds and 72°C for 3 minutes. Another cycle of 94°C for 30 seconds and 70°C for 3 minutes was done proceeded by 94°C for 30 seconds, 68°C for 30 seconds and 72°C for 3 minutes for 36 cycles and a final extension at 72°C for 10 minutes.
Cloning of the PCR product
Extraction of the PCR product was conducted corresponding to Qiagen:QIAquick Gel Extraction Kit and cloned with Invitrogen: TOPO TA Cloning. The clones were selected against the metabolism of X-gal and the production of β–galactosidase purified following the QIAprep spin miniprep kit and sent for sequencing. (Davis Sequencing, Davis, CA)
Embryo collection and maintenance
Mature or maturing Petromyzon marinus adults were obtained from Hammond Bay Biological Station, Millersburg, MI, USA. Mature animals were maintained in our lamprey facility in 12°C chilled recirculating water and used for spawning. Juveniles were kept at lower temperatures before the onset of the maturation and then progressively induced to maturation, by gradually augmenting the water temperature and expanding the daylight cycle. For in vitro fertilization, eggs are stripped manually from a single gravid female into a 500ml crystallizing dish containing 100–200ml of spring water and milt from a spermiated male is then expressed directly onto the eggs. After 15 minutes, the fertilized eggs are washed through several changes of distilled 18°C water and placed in a 4-liter container, in spring water in the 18°C incubator. After the first division the embryos are transferred to 0.1X MMR (Marc’s Modified Ringers) for long-term culture. The medium from each culture is replaced with fresh one every day to avoid fungal infection. Embryos were fixed in MEMFA (4% formaldehyde, 0.1M MOPS (pH 7.4), 1 mM MgSO4, 2 mM EGTA), dehydrated gradually and stored in 100% methanol at −20°C (Sauka-Spengler et al., 2007).
In situ hybridization and histology
Whole-mount in situ hybridization of lamprey embryos was performed using digoxigenin- or RNA probes according to Xu and Wilkinson (Xu et al., 1998), with following modifications: Prior to Proteinase K step, embryos equilibrated in the bleaching solution (0.5X SSC, 5% formamide, 10%H2O2), were exposed to direct light using light box for 10–15 minutes. The concentration and the length of Proteinase K treatment (~20μg/ml, 10 minutes) was the same for embryos of all stages. Hybridization and subsequent washes were carried out at 70°C in hybridization solution containing 50% formamide; 1.3X SSC; 5mM EDTA pH8.0; 200 μg/ml yeast tRNA; 100μg/ml heparin; 0.2% Tween-20 and 0.5% Chaps. The hybridization signal was detected using BM Purple substrate (Roche, Indianapolis, IN) for early stage embryos (E3–E10) or NBT/BCIP (Roche, Indianapolis, IN) for later stages. After photographing, embryos were post-fixed in 4% Paraformaldehyde/PBS, rinsed in PBS, cryo-protected in two subsequent steps: 15%sucrose/PBS and 7.5%gelatin/15% sucrose/PBS, equilibrated and mounted in 20% gelatin/PBS and frozen in liquid nitrogen. 10μm cryosections were collected on Super Frost Plus slides (Fischer Scientific, Pittsburgh, PA).
Phylogenetic Analysis
The amino acid alignments and Neighbor Joining (NJ) tree were constructed using ClustalX. The Maximum Pasimony likelihood tree was build using Mega. The trees were visualized using Tree View v. 0.5.0. Protein sequences from the HMG boxes of Sox family genes were used to build the alignments. Sequences form other species were retrieved from GenBank, and carry the following nomenclature abbreviations: Dr, Danio rerio; Gg, Gallus gallus; Mm, Mus musculus; Pm, Petromyzon marinus; Xl, Xenopus laevis.
Results and Discussion
Phylogenetic Analysis
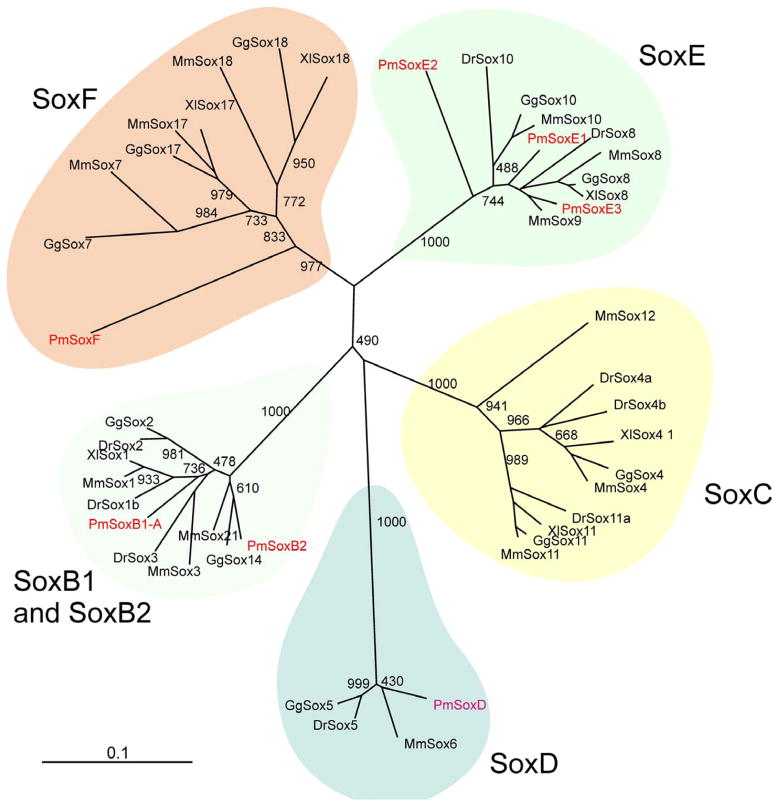
The screening of a full-length cDNA library (Sauka-Spengler et al., 2007) coupled with BLAST searches of the lamprey genome, allowed identification of four new lamprey Sox orthologs, from Sox families B, D E and F. We also included in our analysis the three SoxE orthologs previously described in McCauley et al., 2006.
Three SoxB genes were identified in this study – SoxB1b, SoxB2 and a putative SoxB1a gene. The lamprey SoxB2 groups with the Sox 21/14 in our phylogenetic analysis, while SoxB1-A clusters with the Sox1/2/3 paralogy group. The presence of orthologs of SoxB1 and SoxB2 in the lamprey is expected since the duplication that gave rise to these genes is though to be ancient having occurred before the deuterostome/protostome split (Mckimmie et al., 2005; Zhong et al., 2011). The putative SoxB1a gene we have cloned seems to be an ortholog of the Sox2 gene due to the similarity of the 5′ fragment of the transcript. This gene lacks a HMG box, which precludes a more detailed analysis of its phylogenetic position. However, the expression data obtained for this gene (see bellow) suggests that it is likely related to the other SoxB genes analyzed.
We identified only a single SoxD family in lamprey that is apparently quite divergent from the other vertebrate SoxDs and more similar to Sox6 group. However, this may be an artifact due to the large number of amino acid substitutions observed in this ortholog (Figure 1, S1). The lamprey SoxF, on the other hand, clusters in the base of the Sox7/18/17 branch. Thus, our data suggest that there is one lamprey ortholog for half of the major Sox families (D, F, B2), suggesting that the duplication events that led to the expansion of paralogs in such families took place after the Cyclostome/Gnathostome split.
Figure 1. Phylogenetic Analysis of lamprey Sox genes.
Unrooted phylogenetic tree of Sox genes obtained from a neighbor joining analysis performed on ClustalX. The HMG box protein sequences were used in the construction of the tree. Boxes highlight the families of Sox genes. Abbreviations are Dr, Danio rerio; Gg, Gallus gallus; Mm, Mus musculus; Pm, Petromyzon marinus; Xl, Xenopus laevis.
Expression pattern of SoxB family members
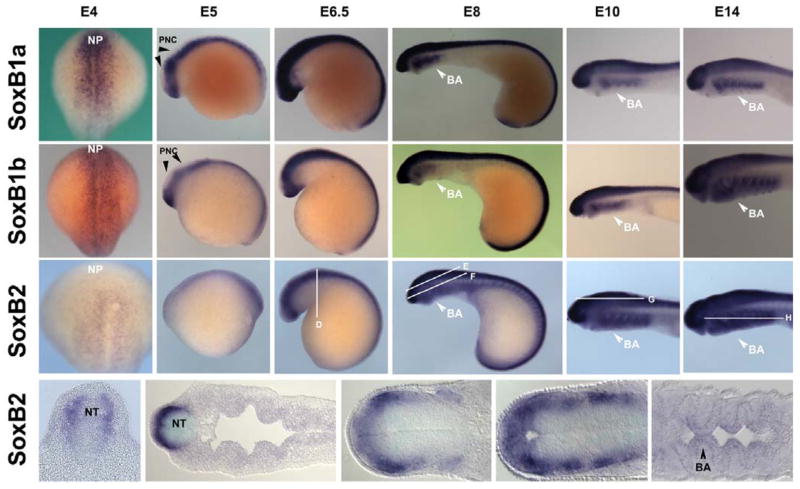
The SoxB family is known to play a major role in neural induction and differentiation. In zebrafish, SoxBs are expressed in an early stage starting at the anterior neural plate and throughout the CNS (Rauch et al., 2003; Thisse et al, 2005). Whole mount in situ hybridization of the SoxB family revealed that SoxB1a, SoxB1b, and SoxB2 are all expressed in the neural plate at embryonic day (E) 4, (Figure 2: A, B & C). Similarly at E5, SoxB1a and SoxB1b have expression patterns in the neural tube with the exception of its anterior-dorsal aspect (Figure 2: A & B), whereas SoxB2 is expressed continuously throughout the dorsal neural tube (Figure 2: C). At E6.5, by which time neural crest cells are within the dorsal aspect of the neural tube preparing to emigrate, SoxB1a and SoxB2 are expressed on the dorsal aspect of the neural tube (Figure 2: A & C) while SoxB1b is absent from the anterior-dorsal region (Figure 2: B). At E8, SoxB genes are expressed in the neural tube as well as in the forming branchial arches. Analysis of sectioned embryos reveals that SoxB2 is expressed mainly in the neural tube, the cranial ganglia, trigeminal ganglia, and in cardiac tissue.
Figure 2. Expression pattern of SoxB genes.
The SoxB genes are expressed within the neural plate, neural tube and branchial arches. SoxB1a, 4–14 days (A): SoxB1a is not expressed in pre-migratory neural crest at E5, with expression initiating at E6 at the onset of neural crest emigration. SoxB1b: 4–14 day (B) SoxB1b is not expressed at E5 or E6 in pre-migratory neural crest. SoxB2; 4–14 day, (C) and sections (D–H) from 6.5 day, 8 day, 10 day, and 14 day, is expressed throughout the neural tube at E5 and E6. Sections reveal SoxB2’s presence at later stages in the neural tube, cranial ganglia, and ectoderm derived portions of the branchial arches. NP: neural plate, PNC: Pre-migratory neural crest, NT: neural tube, BA: branchial arch
Finally between E10–E14, the SoxBs are expressed in the neural tube and in the brachial arches. At E14, SoxB1a and SoxB1b are expressed in the branchial arches. Similarly SoxB2 is expressed in both neural tissue and mesoderm-derived portions of the branchial arches. SoxB2 is also expressed in cardiac and other non-neural crest derived tissue. These data suggest that the SoxB genes are expressed in similar domains but in a distinct temporal sequence, first with SoxB2, followed by SoxB1a and then SoxB1b. Despite their staggered temporal expression, all SoxBs are present in migrating neural crest cells as well as neural crest derivatives such as cranial ganglia and branchial arches. SoxB2 is present in both premigratory and migrating neural crest cells.
In Xenopus, SoxBs play a role in neural plate formation and also have later roles throughout CNS formation (Cunningham et al., 2008; Kishi et al., 2000, Rodgers et al., 2008). In chick, SoxBs are also expressed on the neural plate and along the neural tube. In Mice, SoxBs are all expressed in the primitive streak ectoderm. Sox1 is expressed early on in the neural folds. Sox 2 and 3 are found in the forming neural plate onward (Wood et al., 1999).
Expression pattern of SoxD family members
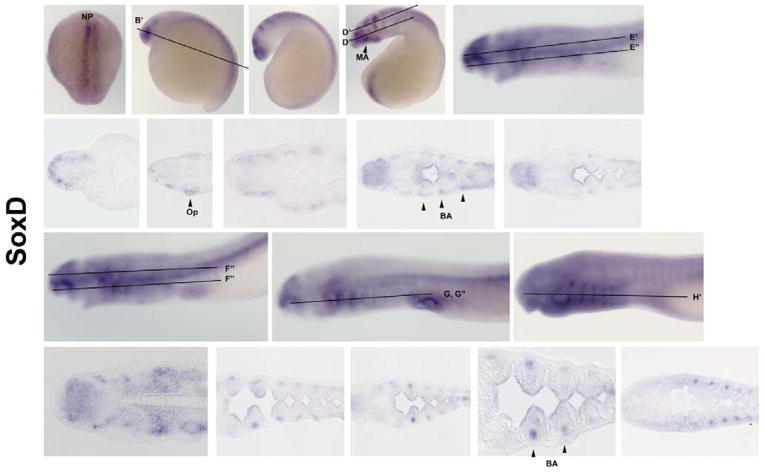
SoxDs are expressed widely in vertebrate neural tissues, forebrain, in fast muscles, somites, mytome, and cardiac precursors (Wang et al., 2011; Von Hofsten et al., 2008; Kudoh et al., 2001). SoxDs are necessary for formation of the notochord and chondrogenesis. They are also found in glial cells and other early NC linages (Smits & Lefebvre, 2003; Lefebvre et al., 1998; Perez-Alcala et al., 2004). In lamprey, SoxD is expressed at high levels on the neural plate border and along the neural folds at E4 (Figure 3: A). At later stages, SoxD is not expressed in the premigratory crest but is expressed along the neural tube (Figure 3: B). At E8, it is observed in the optic vesicle (Figure 3: D & D′). Beginning at E10, SoxD is expressed in the endoderm- and mesoderm-derived portions of the branchial arches (Figure 3: E, E′, & E″). From E12–E16, the mesenchymal portions of the branchial arches condense to form the branchial cartilage (Figure 3: F–H & F″–H′). Interestingly, at E14, SoxD is heavily expressed in the heart but not at prior or subsequent stages.
Figure 3. SoxD Expression.
SoxD is expressed from 4 – 16 day (A–H), sections (B′–H′) beginning on the boarder of the neural plate (A) and progressing to various neural crest derivatives, optic vesicle (D & D′), branchial arches (E, E′ & E″), and branchial arch cartilage (F–H & F″–H′). Beginning E8 (D) SoxD is expressed in forming branchial arch progenitors, optic vesicle and mandibular arch. From E10–E16 (E–H), expression in the forebrain and heart are down decreased over time while expression of SoxD is consistent with branchial arch cartilage condensation. Np: neural plate, Op: optic vesicle, BA: branchial arch. MA: mandibular arch
Expression pattern of SoxE family members
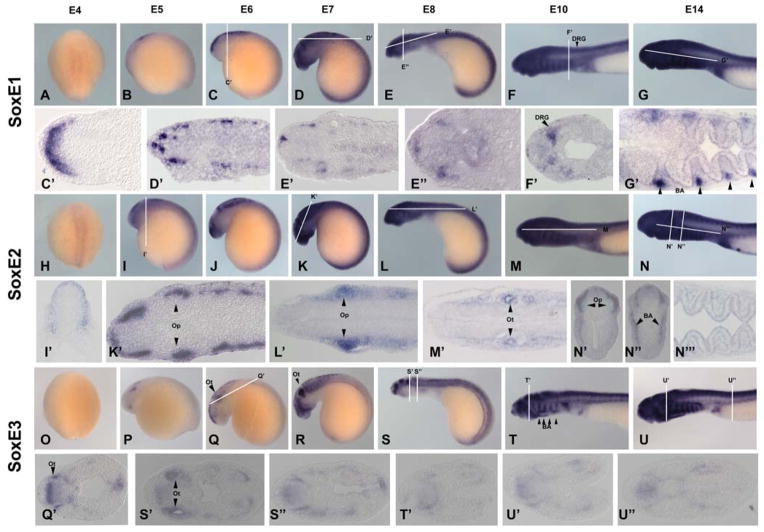
The SoxE family has three subgroups denoted SoxE1, E2, and E3 (McCauley and Bronner-Fraser, 2006). At E4, SoxE1 and SoxE2 both display low levels of expression in the neural plate (Figure 4: A & H) whereas SoxE3 is not yet expressed (Figure 4:O). By E5, all SoxEs exhibit expression in the neural tube as well as distinct domains in the embryo. At E5–E7, SoxE1 is observed in two regions of the anterior-dorsal aspect of the neural tube (Figure 4: B–D). At later stages, it is expressed in the neural tube, cranial ganglia (Figure: 4 D & D′) dorsal root ganglia (Figure 4: F &F′) and in mesoderm and neural crest derived portions of the branchial arches cartilage (Figure 4: G &G′). SoxE2 is expressed similarly but with higher intensity (Figure 4: I–K). At older stages, SoxE2 is expressed in the neural tube, cranial ganglia (Figure 4: K–K′), optic (Figure 4:L & L′), otic vesicles (Figure 4: M & M′), and branchial arches (Figure 4: N–N‴). SoxE3 exhibits a strong signal starting at E5 in the optic vesicle (Figure 4: P). It is strongly expressed during neural crest migration. From E6–E14, SoxE3 is very prominently expressed in the otic vesicles (Figure 4: Q–S, & S′), cranial ganglia (Figure 4: S), neural tube, and branchial arches (Figure 4: S–U & S″–U″).
Figure 4. SoxE Expression.
The SoxE family show low level of early expression with SoxE1 and E2 expression in the neural plate at E4 (A & H). SoxE1 (A–G) begins showing expression in the pre-migratory neural crest (C & C′) then cranial ganglia (D, D′, E, E′, & E″), dorsal root ganglia (F & F′), and the branchial cartilage at late stages (G & G′). SoxE2(H–N) is also expressed heavily in the premigratory neural crest (I & I′) and ultimately in the optic (K, K′, L, L′ and N′), otic vesicles (M&M′), and branchial arches (N–N‴). SoxE3(O–U) is expressed early in the otic vesicle (Q & Q′). It is later expressed heavily in the otic vesicle (S & S′), other crancial ganglia (S–U), neural tube, branchial arches (S–U). Op: optic vesicle, Ot: otic vesicle, BA: branchial arch
Expression pattern of SoxF family member
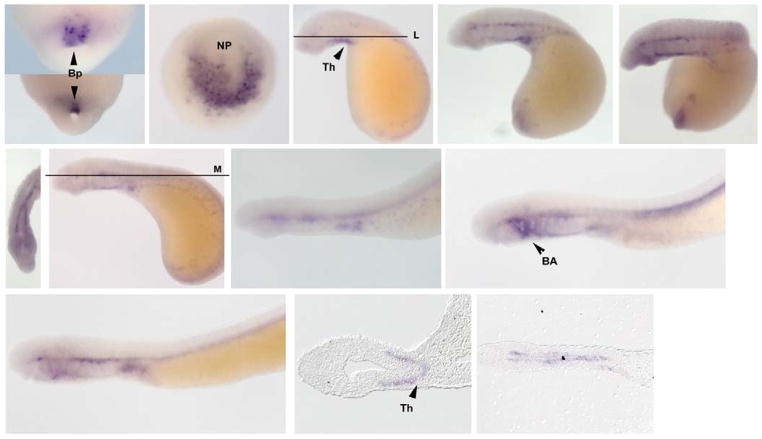
In vertebrates, SoxFs are involved with vascular development and cardiogenesis, are expressed in structures such as the aortic arch, circulatory system, and pharyngeal arches in addition to parts of the CNS (Zhang et al., 2005; Kyuno et al., 2008). We isolated a single SoxF gene. At gastrula stages, SoxF is expressed in involuting cells in the dorsal lip of the blastopore (Figure 5: A & B). At the neurula stage (E4.5), SoxF is found in the neural plate (Figure 5:C). Expression in precursor cells to the thyroid gland begins at E7, whereas by E8, SoxF is observed in tissues surrounding the notochord (Figure 5: D, E and L). Low levels of SoxF are observed in cardiac precursors at E10 (Figure 5: I). At E12, SoxF is more strongly expressed in the anterior branchial arches, similar to that noted for SoxD (Figure 5: J).
Figure 5. SoxF Expression.
SoxF is expressed early within the blastopore [(A & B) ventral view; E4.5] and the neural plate (C dorsal view). Beginning from E7 to E14, SoxF is expressed surrounding the notochord and in precursors to the thyroid, heart, and branchial arches (E–M). Beginning at E7, SoxF begins to express in the forming thyroid (L). Ventral view of E8.5 displaying the forming thyroid (G). In addition, SoxF is temporarily expressed in the first and second branchial arch at E14 and 16 (J and K). Bp: blastopore, NP: neural plate Th: thryroid precursors BA: Branchial Arch
Conclusion
Taken collectively, our phylogenetic and expression data provide interesting insights into the evolution of Sox genes in vertebrates. Surprisingly, we were only able to find one ortholog for half of the major Sox families (D, F, B2). This suggests that either these families expanded after emergence of jawed vertebrates, or that there was extensive loss of these Sox genes in lamprey. For the remaining families, the Sox genes appear to have undergone independent duplications though there may be some bias toward particular paralogs. For example, SoxE1 is most similar to Sox9 and SoxE2 to Sox8. Based on the positions of various genes, we speculate that the SoxB2 gene (Sox14) and SoxD genes either diverged earlier than their gnathostome orthologs or that paralogs were lost, as seen by Sox14 and Sox 5a or 6. SoxF is very similar to all variance of SoxF in jawed vertebrates, suggesting it may be an ancestral gene that was independently duplicated by jawed vertebrates.
Our expression data further support the idea that lamprey genes underwent independent gene duplications, separate from the two rounds of gene duplication observed in other vertebrates. For example, in those Sox families in which we identified multiple members, we find that the lamprey genes often have overlapping expression patterns, the sum of which reflects the overall expression pattern of that Sox family in other vertebrates. Generally, each family has a high degree of overlapping expression. However, no unique ortholog was found with a pattern that is similar only to a single gnathostome Sox gene, except when we found only a single member of that family in lamprey. Sequencing of additional cyclostome genomes will provide clarity as to when such independent duplications may have occurred.
Supplementary Material
Acknowledgments
We thank Natalya Nikitina for her contribution of previously sequenced genes. This work was supported by NIH grant DE017911 to MEB.
Contributor Information
BENJAMIN R. UY, Email: uy.benjamin@gmail.com.
MARCOS SIMOES-COSTA, Email: marcos@caltech.edu.
TATJANA SAUKA-SPENGLER, Email: spengler@caltech.edu.
MARIANNE E. BRONNER, Email: mebronner@gmail.com.
References
- LAUDET V, STEHELIN D, CLEVERS H. Ancestry and diversity of the HMG box superfamily. Nucleic acids research. 1993;21:2493–2501. doi: 10.1093/nar/21.10.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HONG CS, SAINT-JEANNET JP. Sox proteins and neural crest development. Seminars in cell & developmental biology. 2005;16:694–703. doi: 10.1016/j.semcdb.2005.06.005. [DOI] [PubMed] [Google Scholar]
- BETANCUR P, BRONNER-FRASER M, SAUKA-SPENGLER T. Genomic code for Sox10 activation reveals a key regulatory enhancer for cranial neural crest. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3570–3575. doi: 10.1073/pnas.0906596107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHEPERS GE, TEASDALE RD, KOOPMAN P. Twenty pairs of sox: extent, homology, and nomenclature of the mouse and human sox transcription factor gene families. Developmental cell. 2002;3:167–170. doi: 10.1016/s1534-5807(02)00223-x. [DOI] [PubMed] [Google Scholar]
- PRIOR HM, WALTER MA. SOX genes: architects of development. Molecular medicine. 1996;2:405–412. [PMC free article] [PubMed] [Google Scholar]
- JAGER M, QUEINNEC E, HOULISTON E, MANUEL M. Expansion of the SOX gene family predated the emergence of the Bilateria. Molecular phylogenetics and evolution. 2006;39:468–477. doi: 10.1016/j.ympev.2005.12.005. [DOI] [PubMed] [Google Scholar]
- DEHAL P, BOORE JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS biology. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ESCRIVA H, MANZON L, YOUSON J, LAUDET V. Analysis of lamprey and hagfish genes reveals a complex history of gene duplications during early vertebrate evolution. Molecular biology and evolution. 2002;19:1440–1450. doi: 10.1093/oxfordjournals.molbev.a004207. [DOI] [PubMed] [Google Scholar]
- TOMSA JM, LANGELAND JA. Otx expression during lamprey embryogenesis provides insights into the evolution of the vertebrate head and jaw. Developmental biology. 1999;207:26–37. doi: 10.1006/dbio.1998.9163. [DOI] [PubMed] [Google Scholar]
- MCCAULEY DW, BRONNER-FRASER M. Importance of SoxE in neural crest development and the evolution of the pharynx. Nature. 2006;441:750–752. doi: 10.1038/nature04691. [DOI] [PubMed] [Google Scholar]
- NEIDERT AH, VIRUPANNAVAR V, HOOKER GW, LANGELAND JA. Lamprey Dlx genes and early vertebrate evolution. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:1665–1670. doi: 10.1073/pnas.98.4.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHONG L, WANG D, GAN X, YANGM T, HEM S. Parallel expansions of Sox transcription factor group B predating the diversifications of the arthropods and jawed vertebrates. PloS one. 2011;6:e16570. doi: 10.1371/journal.pone.0016570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WRIGHT EM, SNOPEK B, KOOPMAN P. Seven new members of the Sox gene family expressed during mouse development. Nucleic acids research. 1993;21:744. doi: 10.1093/nar/21.3.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOWLES J, SCHEPERS G, KOOPMAN P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Developmental biology. 2000;227:239–255. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- SAUKA-SPENGLER T, BRONNER-FRASER M. A gene regulatory network orchestrates neural crest formation. Nature reviews Molecular cell biology. 2008;9:557–568. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- MORALES AV, PEREZ-ALCALA S, BARBAS JA. Dynamic Sox5 protein expression during cranial ganglia development. Developmental dynamics: an official publication of the American Association of Anatomists. 2007;236:2702–2707. doi: 10.1002/dvdy.21282. [DOI] [PubMed] [Google Scholar]
- WAKAMATSU Y, ENDO Y, OSUMI N, WESTON JA. Multiple roles of Sox2, an HMG-box transcription factor in avian neural crest development. Developmental dynamics: an official publication of the American Association of Anatomists. 2004;229:74–86. doi: 10.1002/dvdy.10498. [DOI] [PubMed] [Google Scholar]
- SAUKA-SPENGLER T, MEULEMANS D, JONES M, BRONNER-FRASER M. Ancient evolutionary origin of the neural crest gene regulatory network. Developmental cell. 2007;13:405–420. doi: 10.1016/j.devcel.2007.08.005. [DOI] [PubMed] [Google Scholar]
- XU Q, WILKINSON DG. In situ hybridisation of mRNA with hapten labelled probes. In: Wilkinson DG, editor. In Situ Hybridisation: A Practical Approach. Oxford: Oxford Press; 1998. [Google Scholar]
- MCKIMMIE C, WOERFEL G, RUSSELL S. Conserved genomic organisation of Group B Sox genes in insects. BMC genetics. 2005;6:26. doi: 10.1186/1471-2156-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAUCH GJ, LYONS DA, MIDDENDORF I, FRIEDLANDER B, ARANA N, REYES T, TALBOT WS. Submission and Curation of Gene Expression Data. ZFIN Direct Data Submission. 2003 ( http://zfin.org)
- THISSE B, PFLUMIO S, FÜRTHAUER M, LOPPIN B, HEYER V, DEGRAVE A, WOEHL R, LUX A, STEFFAN T, CHARBONNIER XQ, THISSE C. Expression of the zebrafish genome during embryogenesis (NIH R01 RR15402) ZFIN Direct Data Submission. 2001 ( http://zfin.org)
- WANG X, ONO Y, TAN SC, CHAI RJ, PARKIN C, INGHAM PW. Prdm1a and miR-499 act sequentially to restrict Sox6 activity to the fast-twitch muscle lineage in the zebrafish embryo. Development. 2011;138:4399–4404. doi: 10.1242/dev.070516. [DOI] [PubMed] [Google Scholar]
- VON HOFSTEN J, ELWORTHY S, GILCHRIST MJ, SMITH JC, WARDLE FC, INGHAM PW. Prdm1- and Sox6-mediated transcriptional repression specifies muscle fibre type in the zebrafish embryo. EMBO Rep. 2008;9(7):683–689. doi: 10.1038/embor.2008.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KUDOH T, TSANG M, HUKRIEDE NA, CHEN X, DEDEKIAN M, CLARKE CJ, KIANG A, SCHULTZ S, EPSTEIN JA, TOYAMA R, DAWID IB. A gene expression screen in zebrafish embryogenesis. ZFIN Direct Data Submission. 2001 doi: 10.1101/gr.209601. ( http://zfin.org) [DOI] [PubMed]
- PENDEVILLE H, WINANDY M, MANFROID I, NIVELLES O, MOTTE P, PASQUE V, PEERS B, STRUMAN I, MARTIAL JA, VOZ ML. Zebrafish Sox7 and Sox18 function together to control arterial-venous identity. Dev Biol 2008. 2008 May 15;317(2):405–16. doi: 10.1016/j.ydbio.2008.01.028. Epub 2008 Feb 7. [DOI] [PubMed] [Google Scholar]
- THISSE B, THISSE C. Fast Release Clones: A High Throughput Expression Analysis. ZFIN Direct Data Submission. 2004 ( http://zfin.org)
- ROGERS CD, HARAFUJI N, ARCHER T, CUNNINGHAM DD, CASEY ES. Xenopus Sox3 activates sox2 and geminin and indirectly represses Xvent2 expression to induce neural progenitor formation at the expense of non-neural ectodermal derivatives. Mech Dev. 2009;126:42–55. doi: 10.1016/j.mod.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KISHI M, MIZUSEKI K, SASAI N, YAMAZAKI H, SHIOTA K, NAKANISHI S, SASAI Y. Requirement of Sox2-mediated signaling for differentiation of early Xenopus neuroectoderm. Development. 2000;127:791–800. doi: 10.1242/dev.127.4.791. [DOI] [PubMed] [Google Scholar]
- Cunningham DD, Meng Z, Fritzsch B, Casey ES. Cloning and developmental expression of the soxB2 genes, sox14 and sox21, during Xenopus laevis embryogenesis. Int J Dev Biol 2008. 2008;52(7):999–1004. doi: 10.1387/ijdb.082586dc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARTIN BL, HARLAND RM. Hypaxial muscle migration during primary myogenesis in Xenopus laevis. Dev Biol. 2001 Nov 15;239(2):270–80. doi: 10.1006/dbio.2001.0434. [DOI] [PubMed] [Google Scholar]
- ZHANG C, BASTA T, KLYMKOWSKY MW. SOX7 and SOX18 are essential for cardiogenesis in Xenopus. Dev Dyn 2005. 2005 Dec;234(4):878–891. doi: 10.1002/dvdy.20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KYUNO J, MASSÉ K, JONES EA. A functional screen for genes involved in Xenopus pronephros development. Mech Dev. 2008 Jul 01;125(7):571–586. doi: 10.1016/j.mod.2008.03.001. [DOI] [PubMed] [Google Scholar]
- REX M, ORME A, UWANOGHO D, TOINTON K, WIGMORE PM, SHARPE PT, SCOTTING PJ. Dynamic expression of chicken Sox2 and Sox3 genes in ectoderm induced to form neural tissue. Dev Dyn. 1997 Jul;209(3):323–332. doi: 10.1002/(SICI)1097-0177(199707)209:3<323::AID-AJA7>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- PEREZ-ALCALA S, NIETO MA, BARBAS JA. LSox5 regulates RhoB expression in the neural tube and promotes generation of the neural crest. Development. 2004;131:4455–4465. doi: 10.1242/dev.01329. [DOI] [PubMed] [Google Scholar]
- JAVERZAT S, FRANCO M, HERBERT J, PLATONOVA N, PEILLE AL, PANTESCO V, DE VOS J, ASSOU S, BICKNELL R, BIKFALVI A, HAGEDORN M. Correlating global gene regulation to angiogenesis in the developing chick extra-embryonic vascular system. PLoS One. 2009 Nov 17;4(11):e7856. doi: 10.1371/journal.pone.0007856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WOOD HB, EPISKOPOU V. Comparative expression of the mouse Sox1, Sox2 and Sox3 genes from pre-gastrulation to early somite stages. Gene expression pattern. 1999 Aug;86(1–2):197–201. doi: 10.1016/s0925-4773(99)00116-1. [DOI] [PubMed] [Google Scholar]
- SMITS P, LEFEBVRE V. Sox5 and Sox6 are required for notochord extracellular matrix sheath formation, notochord cell survival and development of the nucleus pulposus of intervertebral discs. Development. 2003;130:1135–1148. doi: 10.1242/dev.00331. [DOI] [PubMed] [Google Scholar]
- LEFEBVRE V, LI P, DE CROMBRUGGHE B. A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. Embo J. 1998;17:5718–5733. doi: 10.1093/emboj/17.19.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEREZ-ALCALA S, NIETO MA, BARBAS JA. LSox5 regulates RhoB expression in the neural tube and promotes generation of the neural crest. Development. 2004;131:4455–4465. doi: 10.1242/dev.01329. [DOI] [PubMed] [Google Scholar]
- PFISTER S, JONES VJ, POWER M, TRUISI GL, KHOO PL, STEINER KA, KANAI-AZUMA M, KANAI Y, TAM PP, LOEBEL DA. Sox17-dependent gene expression and early heart and gut development in Sox17-deficient mouse embryos. Int J Dev Biol. 2011;55(1):45–58. doi: 10.1387/ijdb.103158sp. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.