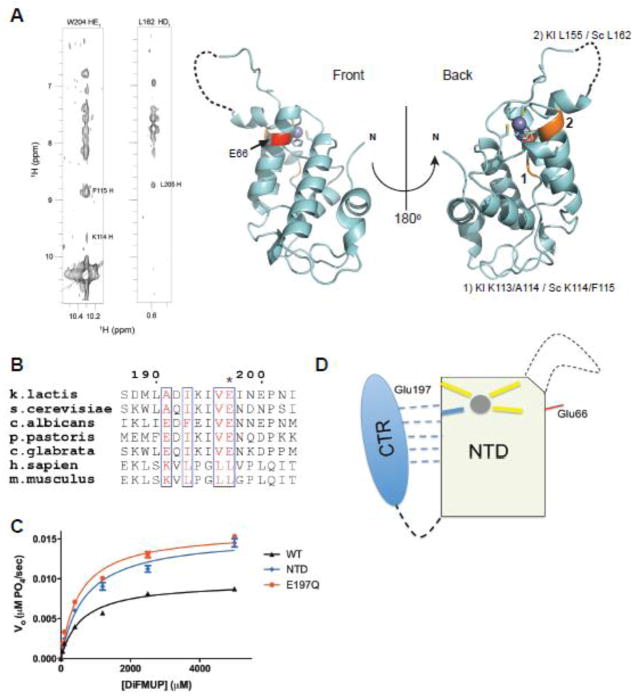
Figure 3. Yeast Rtr1 proteins are auto-inhibited by the C-terminal region.
a) NOE connectivities between residues in the CTR of ScRtr1 to residues in the NTD (left panel). The equivalent residues within the NTD that the CTR makes contacts with are highlighted in orange on the crystal structure of the K. lactis protein, and indicated in numbers. The invariant Glu66 residue is highlighted in red.
b) Sequence alignment of Rtr1 (numbering based on K. lactis) across yeasts and vertebrates, focusing on the extreme C-terminus of the protein. Glu197 is asterisked.
c) Steady state phosphatase assay (n = 3) against DiFMUP of KlRtr1 E197Q (red) compared with full-length protein (black) and with the NTD phosphatase domain (blue). Curves for full-length and NTD are derived from Fig. 1C and Fig. 2B respectively.
d) Cartoon illustration of the regulation of Rtr1 activity by its C-terminus. Zinc is shown as a grey sphere and the structurally dynamic loop is shown as a dashed line. Glu66 is denoted by a red line on the same face as the active site forming loop. The CTR weakly interacts with the NTD in the absence of a metal, as shown by the dashed lines; among interacting residues is the critical Glu197 necessary for regulation.