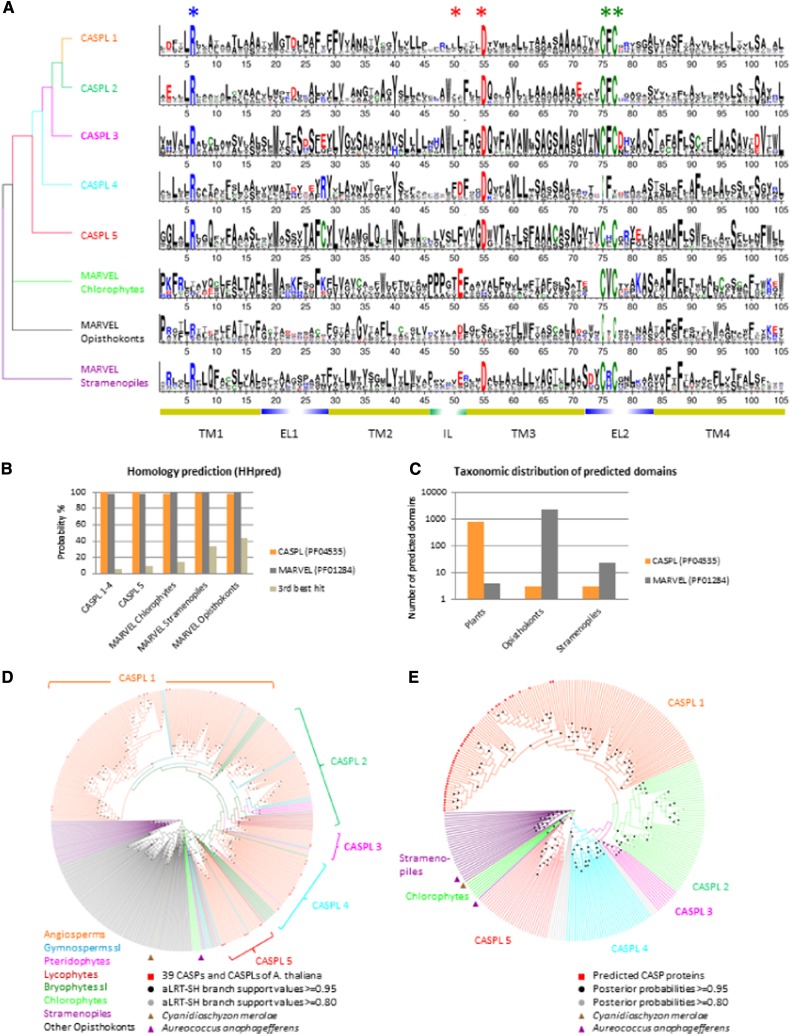
Figure 1.
Evidence for a common origin of CASPLs and MARVELs. A, Sequence similarity and five conserved residues. Sequence logos covering the four transmembrane domains and conserved adjacent regions of the CASPL/MARVEL domains have been constructed for the five plant groups, as well as for chlorophytes, stramenopiles, and opisthokonts. Conserved residues are marked by asterisks. Yellow bars below logos correspond to the predicted transmembrane domains of CASP1 from Arabidopsis, and the adjacent conserved regions of the assumed extracellular loops (EL1 and EL2) and of the intracellular loop (IL) are indicated. The predicted relationship of the five CASPL groups and MARVEL taxonomic groups is shown as a tree at the left of the sequence logos. The color code of the protein groups corresponds to that in E. B, Homology prediction. CASPL and MARVEL domains are predicted to belong to both domain families with a probability of at least 97.3% (Supplemental Table S1). C, Taxonomic range for domain predictions. A total of 99.5% of the CASPL domains are predicted in plant genes, and 99.9% of the MARVEL domains are predicted in genes from species of the animal lineage. D, Circular cladogram based on the maximum likelihood phylogeny of the CASPL and MARVEL domains. Genes from species in key positions of the CASPL/MARVEL domain phylogeny with no stable position in the tree are marked by triangles: two genes from the stramenopiles Aureococcus anophagefferens (UniProtKB nos. F0YFA3 and F0Y3N5) and one gene from the rhodophyte Cyanidioschyzon merolae (UniProtKB no. M1VAW8). Branch colors indicate taxonomic species groups. E, Circular cladogram of the CASPL phylogeny with MARVELs from stramenopiles and algae as an outgroup, based on a Bayesian inference analysis. The five CASPL subfamilies are marked by colors, and stramenopiles and chlorophytes are colored as in D; unclassified genes are gray. [See online article for color version of this figure.]