Abstract
The Genetic Investigation of Anthropometric Traits (GIANT) consortium identified 14 loci in European Ancestry (EA) individuals associated with waist-to-hip ratio (WHR) adjusted for body mass index. These loci are wide and narrowing the signals remains necessary. Twelve of 14 loci identified in GIANT EA samples retained strong associations with WHR in our joint EA/individuals of African Ancestry (AA) analysis (log-Bayes factor >6.1). Trans-ethnic analyses at five loci (TBX15-WARS2, LYPLAL1, ADAMTS9, LY86 and ITPR2-SSPN) substantially narrowed the signals to smaller sets of variants, some of which are in regions that have evidence of regulatory activity. By leveraging varying linkage disequilibrium structures across different populations, single-nucleotide polymorphisms (SNPs) with strong signals and narrower credible sets from trans-ethnic meta-analysis of central obesity provide more precise localizations of potential functional variants and suggest a possible regulatory role. Meta-analysis results for WHR were obtained from 77 167 EA participants from GIANT and 23 564 AA participants from the African Ancestry Anthropometry Genetics Consortium. For fine mapping we interrogated SNPs within ±250 kb flanking regions of 14 previously reported index SNPs from loci discovered in EA populations by performing trans-ethnic meta-analysis of results from the EA and AA meta-analyses. We applied a Bayesian approach that leverages allelic heterogeneity across populations to combine meta-analysis results and aids in fine-mapping shared variants at these locations. We annotated variants using information from the ENCODE Consortium and Roadmap Epigenomics Project to prioritize variants for possible functionality.
INTRODUCTION
Waist–hip ratio, a measure of body fat distribution, is associated with metabolic consequences independent of overall adiposity as measured by body mass index (BMI) (1–3). Evidence has indicated that body fat distribution is partially determined by genetic factors with age- and BMI-adjusted heritability estimates for waist–hip ratio ranging from 36–61% (4).
In the past few years, genome-wide association studies have seen numerous successes in the identification of genetic variants associated with adiposity traits, including those characterizing centralized fat patterning. The Genetic Investigation of Anthropometric Traits (GIANT) consortium previously reported 14 loci associated with waist–hip ratio adjusted for BMI, age, age2 and sex [waist-to-hip ratio (WHR)] in studies of European Ancestry (EA) (5). Recently, we also conducted a similar genome-wide association analysis in African Ancestry (AA) studies jointly from the African Ancestry Anthropometry Genetics (AAAG) Consortium, identifying one WHR-associated locus (6). While our analysis failed to detect genome-wide significant association findings overlapping those in the GIANT Consortium, our lead single-nucleotide polymorphism (SNP) rs6931262 at newly identified RREB1 in AA is 474 kb away from the lead SNP rs1294421 at LY86 in EA. Given the low pairwise linkage disequilibrium (LD), these variants likely represent two independent signals. However, they may be also partially tagging an untyped functional variant that contributes to both underlying associations. In addition, 12 of 14 SNPs had the same effect direction with respect to the beta coefficient (P-value = 0.0065) between AA and EA samples and five index SNPs in EA demonstrated nominal significance (P-value < 0.05) in AA. These results may demonstrate similarity in the genetic architecture in EA and AA and suggests that trans-ethnic association analysis may provide further information in fine-mapping the previously identified loci.
Most previous genome-wide association studies (GWAS) have been conducted separately by race/ethnicity due to concerns with allelic heterogeneity and differing patterns of LD between populations. In the present study, we use a Bayesian approach to exploit precisely these differences between EA and AA samples to fine map variants at WHR loci first identified in EA samples.
RESULTS
MANTRA results of 14 previously identified GIANT loci
Twelve of 14 previously published loci (index SNP) in the samples of EA retained strong evidence for association (logBF > 6.1) with WHR in the joint trans-ethnic analysis of EA and AA samples (Table 1). We used a stringent threshold, i.e. logBF value of at least 6.1 based on empirical simulation results reported by Wang et al. (7), which most closely approximates genome-wide significance. None of the 14 loci displayed heterogeneity in their allelic effects (all posterior probabilities for heterogeneity <0.95). At four loci, the originally reported associated SNP remained the lead SNP for that region (i.e. the SNP with the largest logBF within each locus): rs984222 at TBX15-WARS2; rs6784615 at NISCH-STAB1; rs1443512 at HOXC13; and rs4823006 at ZNRF3-KREMEN1. For the remaining 10 loci, different SNPs with greater effect sizes and reduced credible regions were identified; all alternate SNPs are in LD with the previously identified variants in EA (r2 > 0.4 in HapMap II CEU).
Table 1.
Trans-ethnic meta-analysis association results for 14 loci previously reported in EA sample
| Loci information |
EA-sample only | Multi-ethnic meta-analysis results | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genes | Index SNPa | chr | LogBFb | Lengthc | nd | Lead SNP | LogBFe | Lengthc | nd | CEU LDf | YRI LDf |
| TBX15-WARS2 | rs984222 | 1 | 12.68 | 27 654 | 4 | rs984222 | 14.69 | 1591 | 3 | Same | Same |
| DNM3-PIGC | rs1011731 | 1 | 8.95 | 54 037 | 11 | rs9286854 | 10.01 | 48 775 | 10 | 0.9 (1.0) | 0.5 (0.8) |
| LYPLAL1 | rs4846567 | 1 | 10.28 | 14 8141 | 23 | rs2820443 | 11.84 | 27 409 | 8 | 1.0 (1.0) | 0.9 (1.0) |
| GRB14 | rs10195252 | 2 | 8.60 | 31 272 | 4 | rs1128249 | 12.10 | 31 272 | 5 | 0.9 (1.0) | 1.0 (1.0) |
| NISCH-STAB1 | rs6784615 | 3 | 5.83 | 27 7969 | 10 | rs6784615 | 5.61 | 43 1135 | 10 | Same | Same |
| ADAMTS9 | rs6795735 | 3 | 5.78 | 30 676 | 13 | rs4132228 | 8.48 | 8669 | 7 | 0.4 (1.0) | 0.1 (1.0) |
| CPEB4 | rs6861681 | 5 | 5.03 | 93 159 | 11 | rs10516107 | 5.47 | 93 159 | 12 | 1.0 (1.0) | N/A |
| LY86 | rs1294421 | 6 | 7.41 | 15 862 | 6 | rs1294410 | 9.89 | 6952 | 5 | 0.8 (1.0) | 0.4 (0.7) |
| VEGFA | rs6905288 | 6 | 8.43 | 5678 | 2 | rs1358980 | 10.83 | 6655 | 2 | 0.6 (0.9) | 0.1 (1.0) |
| RSPO3 | rs9491696 | 6 | 12.89 | 66 820 | 33 | rs7766106 | 16.42 | 60 186 | 20 | 1.0 (1.0) | 0.8 (0.9) |
| NFE2L3 | rs1055144 | 7 | 7.03 | 38 160 | 13 | rs4141278 | 6.91 | 36 707 | 10 | 1.0 (1.0) | 0.0 (1.0) |
| ITPR2-SSPN | rs718314 | 12 | 6.96 | 38 192 | 13 | rs7302344 | 8.25 | 28 393 | 8 | 0.6 (0.9) | 0.1 (1.0) |
| HOXC13 | rs1443512 | 12 | 6.71 | 7447 | 3 | rs1443512 | 7.27 | 7447 | 3 | Same | Same |
| ZNRF3-KREMEN1 | rs4823006 | 22 | 6.55 | 2194 | 3 | rs4823006 | 6.84 | 2194 | 2 | Same | Same |
aTop variants identified in Heid et al. (2010) (5).
bEvidence of association for the index SNP in EA-sample only.
cLength of 95% credible region in base pair.
dThe number of SNPs of interest, potential causal or tagging to the causal, within the region.
eEvidence of association for the lead SNP in EA + AA sample.
fLD information, r2 (D′), between the index SNP and the lead SNP.
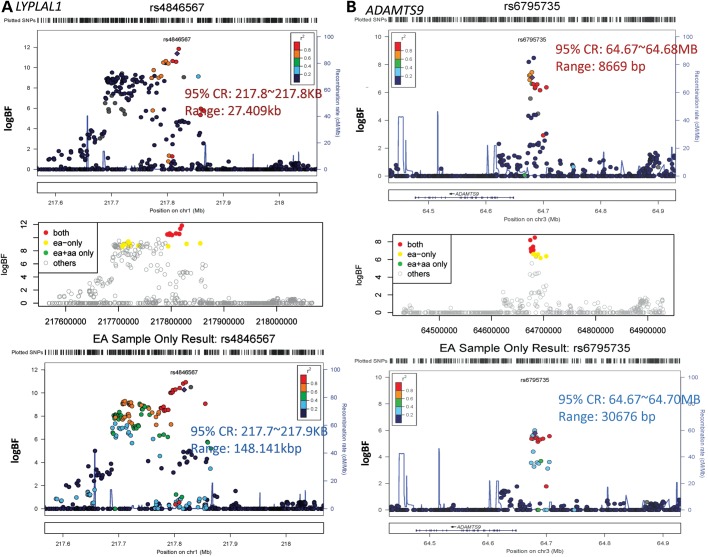
We constructed 95% credible sets (CSs) for the meta-analysis results from GIANT EA samples only and for the trans-ethnic meta-analysis result (Table 1). Eight of the 95% CSs obtained from trans-ethnic association analyses generated shorter CSs length ranging from 4 to 94% length reduction (DNM3-PIGC, NFE2L3, LY86, LYPLAL1, ADAMTS9, ITPR2-SSPN, RSPO3 and TBX15-WARS2) compared with the sets based on EA samples only (Fig. 1 and Supplementary Material, Fig. S1) and five (LY86, LYPLAL1, ADAMTS9, ITPR2-SSPN and TBX15-WARS2) of them have >25% length reduction. The greatest decrease in the distance spanned by the 95% CS of SNPs was at the locus LYPLA1, where the CS was reduced by ∼80% from 148 141 bp in the analysis of EA samples only to 27 409 bp in the trans-ethnic analysis (Fig. 1A). The LD surrounding the original index SNP in this locus is much stronger in CEU compared with that in YRI. In addition, a few SNPs in high LD with the index SNP in both populations have enhanced association signals and the LD block with the index SNP is much narrower in YRI samples, leading to a much narrower CS in trans-ethnic analysis and dropping 15 variants from EA-derived CS. The distance spanned by SNPs at the locus ADAMTS9 decreased 72% from 30 679 to 8669 bp by comparing trans-ethnic analysis to the EA sample only analysis (Fig. 1B). Neither CEU nor YRI have strong LD with many variants surrounding the original index SNPs; however, the variants in strong LD with the index SNP are within a narrow region in both samples of ancestries (narrower in YRI than in CEU). The association signals are highly enhanced and association signals for those top variants are more distinguishable in trans-ethnic analysis compared with EA sample only analysis. This result leads to dropping six variants from EA-derived CS to form a more compact and narrower trans-ethnic-derived CS. Compared with EA results, 4 of 14 CSs (GRB14, CPEB4, HOXC13 and ZNRF3-KREMEN1) remain the same length, and 2 of 14 loci (NISCH-STAB1 and VEGFA) had longer credible regions. Among these six loci, only one CS (NISCH-STAB1) has a substantial increase in the length of credible regions (Supplementary Material, Fig. S1). At this locus there is very weak LD surrounding the original index SNP in EA sample. In addition, the signals for the index SNP and most variants in the region are weak in both EA and AA samples and the trans-ethnic analysis does not enhance the association signals (logBF < 6.1). Therefore, the CS in this case may not be informative.
Figure 1.
Regional plot of loci LYPLAL1 and ADAMTS9. The top panel is obtained from the trans-ethnic meta-analysis result with HapMAP II YRI LD information. The middle panel classifies variants based on whether they are included in the none, either or both of CSs. The red points represent variants retained in both credible regions constructed using EA and trans-ethnic samples; yellow and green points represent variants retained in CSs constructed using EA and trans-ethnic samples, separately; the gray points represents variants which do not fall in any CS.The bottom panel is from EA sample only analysis with HapMap II CEU linkage disequilbrium information. (A) Regional plot of loci LYPLAL1 (Chr1:217 413 815–217 452 830). There are eight SNPs commonly shared by the CSs obtained from EA-only and EA + AA. Fifteen additional SNPs are included in the EA-only derived CS while no additional SNP is included in the EA + AA-derived CS. The length change from 148 141 to 27 409 bp for EA-only and EA + AA derived CSs, respectively. The LD surrounding the original index SNP is much stronger in CEU compared with that in YRI. In addition, few SNPs with high LD with the index SNP in both populations have enhanced association signals and the LD block with the index SNP is much narrower in YRI samples, leading to a much narrower CS in trans-ethnic analysis and dropping 15 variants from EA-derived CS. (B). Regional plot of loci ADAMTS9 (Chr3:64 575 591–64 648 405). There are 7 SNPs commonly shared by the CSs obtained from EA-only and EA + AA. Six additional SNPs are included in the EA-only derived CS while no additional SNP is included in the EA + AA-derived CS. The length change from 30 676 to 8669 bp for EA-only and EA + AA derived CSs, respectively. Neither CEU nor YRI have strong LD for many variants surrounding the original index SNPs; however, the variants with strong LD with index SNP are within a narrow region in both samples of ancestries (narrower in YRI than in CEU). The association signals are highly enhanced and association signals for those top variants are more distinguishable in trans-ethnic analysis compared with EA sample only analysis. These lead to the dropping of six variants from EA-derived CS to form a more compact and narrower trans-ethnic analysis derived CS.
Bioinformatic annotations for the SNPs in CSs
To evaluate the regulatory potential of the CS SNPs at the five loci with the greatest decrease (>25%) in CS length, we examined whether these SNPs map within candidate regulatory elements identified by the ENCODE Consortium and Roadmap Epigenomics Project. We analyzed regulatory elements defined as experimentally detected regions of open chromatin, histone modification enrichment, and transcription factor binding in tissues (blood, brain, endothelial, liver, muscle and pancreatic islet in ENCODE Consortium; adipose, brain, liver, muscle and pancreatic islet in Roadmap Epigenomics Project) that we hypothesize may play a role in WHR pathways (Supplementary Material, Table S1). In total, 11 of the 32 (34.4%) CS SNPs at these loci overlapped elements in two or more datasets in the same tissue, suggesting that they are located in regions with evidence of regulatory activity. The SNPs overlapped regulatory elements from an average of 11 datasets and a maximum of 23 datasets.
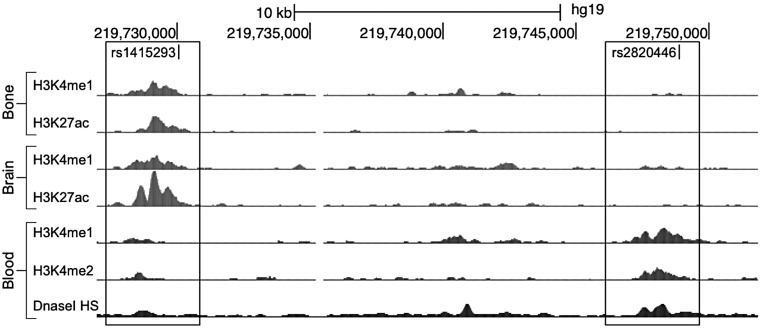
Two SNPs at LYPLAL1 are located in regions with evidence of regulatory activity in distinct tissues (Table 2). While rs1415293 maps within regulatory elements in bone and neuronal tissues, rs2820446 is in a region of regulatory elements in blood (Fig. 2). At the TBX15-WARS2 locus we identified three SNPs within a 1.6 kb region with strong evidence of regulatory activity in adipose, bone, muscle and liver tissues (Table 2). At ITPR2-SSPN, rs7132434 overlaps the largest number (23) of regulatory element datasets.
Table 2.
CS SNPs overlapping evidence of regulatory elements
| CS SNPs |
Regulatory datasets overlapping SNPs (by tissue) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Loci/SNP | Chr | Position | Nearest coding TSS |
Na | Open chromatinb | H3K4me1 | H3K27ac | H3K4me3 | H3K9ac | H3K4me2 | |
| TBX15-WARS2 | |||||||||||
| rs984222 | 1 | 119 503 843 | 26 585 | TBX15 | 19 | AOMLB | AOML | OL | L | OL | |
| rs984225 | 1 | 119 504 284 | 26 144 | TBX15 | 16 | AOML | AOML | OL | L | OL | |
| rs10923712 | 1 | 119 505 434 | 24 994 | TBX15 | 10 | AOML | AOML | L | |||
| LYPLAL1 | |||||||||||
| rs1415293 | 1 | 219 730 006 | 371 938 | SLC30A10 | 4 | ON | ON | ||||
| rs2820446 | 1 | 219 748 818 | 353 126 | SLC30A10 | 3 | B | B | B | |||
| ADAMTS9 | |||||||||||
| rs4504165 | 3 | 64 701 890 | −28 214 | ADAMTS9 | 4 | AM | M | ||||
| LY86 | |||||||||||
| rs912056 | 6 | 6 736 197 | 147 270 | LY86 | 5 | AM | M | M | |||
| rs1294407 | 6 | 6 738 103 | 149 176 | LY86 | 14 | AOM | AM | AOM | OME | ||
| rs1294409 | 6 | 6 738 355 | 149 428 | LY86 | 9 | AM | A | AOM | OM | ||
| ITPR2-SSPN | |||||||||||
| rs7132434 | 12 | 26 472 562 | 123 957 | SSPN | 23 | MNILB | AOMNELB | AOMN | M | OML | |
| rs1049376 | 12 | 26 491 475 | 142 870 | SSPN | 14 | MNE | MN | N | MN | M | |
For loci with >25% decrease in CS size, CS SNPs overlapping two or more regulatory datasets in the same tissue are shown. Negative distance from nearest GENCODE v12 basic annotation TSS indicates the variant is downstream of the TSS relative to the direction of transcription. Tissues with elements overlapping each SNP are indicated as A, adipose; B, blood; E, endothelial; I, pancreatic islets; L, liver; M, muscle; N, brain; O, bone; Chr, chromosome; TSS, transcription start site.
aNumber indicates the total number of overlapping datasets across experiments and cell types.
bOverlap with FAIRE and/or DNaseI hypersensitivity elements indicates open chromatin.
Figure 2.
CS SNPs at LYPLAL1 in regions with evidence of regulatory activity in distinct tissues UCSC genome browser signal enrichment tracks from regulatory datasets with elements overlapping rs1415293 (left-most box; bone and brain) and rs2820446 (right-most box; blood) are shown.
We also examined variants in LD (r2 > 0.8) with any of the thirty-two 95% CS SNPs in either the European (EUR) or African (AFR) 1000 Genomes samples. Ten additional variants overlap regions with evidence of regulatory activity (Supplementary Material, Table S2), usually from the same tissues as the CS SNPs at the locus.
DISCUSSION
We performed association analysis of WHR at 14 previously published loci via trans-ethnic meta-analysis. Among the 14 loci, 9 of the 95% CSs obtained from trans-ethnic association studies contained fewer SNPs. Additionally, the trans-ethnic association studies shortened the length of the 95% credible regions for eight loci, and five of them shortened the length >25%: TBX15-WARS2 (from 27 654 to 1591 bp), ADAMTS9 (from 30 676 to 8669 bp), LY86 (from 15 862 to 6952 bp), LYPLAL1 (from 148 141 to 27 409 bp) and ITPR2-SSPN (from 38 192 to 28 393 bp).
We observed that several of the loci had a different lead SNP in the trans-ethnic results compared with the EA only result. It is likely that when a signal is very strong in a locus and there are few variants in the locus that are in high LD, the lead SNP in trans-ethnic analysis will remain the same. An explanation for the lead SNP changing is LD differences between EA and AA samples. The best functional variant-tagged SNP may differ due to the varying LD structure across different populations and the trans-ethnic meta-analysis accounts for allelic heterogeneity that may lead to a different strongest associated SNP. In addition, the association evidence for a newly identified best SNP is often not much different from the evidence of original index SNP, so random fluctuation may also contribute to the lead SNP changing.
The structure of LD and the strength of signals within each locus likely influence the length of a CS. We observe that some CSs become substantially reduced in size in trans-ethnic meta-analysis results compared their counterpart in EA-only meta-analysis in the loci such as LYPLAL1, ADAMTS9 and LY86. These loci share similar patterns in that there is a strong signal in the EA sample and the signal becomes enhanced in the multi-ethnic meta-analysis. In addition, the LD is weaker in the AA sample within these loci, thus highlighting several top variants. These properties lead to reduced CSs in trans-ethnic meta-analysis. On the other hand, some loci do not have a strong enough signal in the EA sample and weak LD pattern within the locus, such as NISCH-STAB1, leading to uninformative CSs (Supplementary Material, Fig. S1).
Many genetic association signals are shared across populations. Ioannidis et al. (8) demonstrated that for common variants, the magnitude of effect estimates was similar across racial groups; however, allele frequencies varied across racial groups, resulting in differences in the power to detect effects across populations. Several studies examining a range of traits have reported the transferability of disease susceptible genetic loci across different race/ethnic populations (6,9,10). There have been several trans-ethnic analyses reported (11–13); however, there is no analysis, to our knowledge, reporting on the genetic associations for central adiposity by jointly analyzing data from different race/ethnic populations simultaneously. Therefore, assuming that there is a shared signal across populations, our analysis employing MANTRA offers additional insight to uncover the genetic architecture for WHR by taking advantage of results from different race/ethnic groups.
Examining overlap with regulatory elements narrowed a list of 86 candidate SNPs at five loci to 21 SNPs that may influence transcription in WHR relevant cell lines and tissues. LYPLAL1 SNPs overlap two regions with evidence of regulatory activity in different tissues, suggesting that SNPs in these regions may have distinct influences on transcription in different tissues. Further testing is needed to identify whether these variants influence transcriptional activity in these cell types. We also identified a 1.6 kb region near TBX15-WARS2 with evidence of regulatory activity in multiple tissues including adipose tissue. One SNP in this region, rs984222, has been previously reported to have cis-regulatory effects on TBX15 transcription in omental adipose tissue (5), further supporting the plausibility that this or another nearby SNP may influence transcriptional activity. A region near the transcription start site (<100 bp) of the lncRNA, RP11-513G19.1, overlaps more regulatory elements than any other CS SNP evaluated and has strong evidence of enhancer activity in most of the tissues tested. The epigenomic data do not distinguish whether this candidate regulatory element influences transcription of the lncRNA, or a more distal gene such as ITPR2 or SSPN. Further studies are needed to elucidate the regulatory effect of these SNPs on nearby transcripts and their connections to WHR biology.
This analysis identifies CS SNPs overlapping regions with evidence of regulatory activity that suggest good candidates for follow-up studies and also provides insight into the possible tissues in which these variants may regulate transcription. Including additional regulatory datasets from these or other tissues may identify additional candidate regulatory variants and target tissues.
In summary, we performed a trans-ethnic meta-analysis for WHR with previously published EA and AA meta-analysis results using an analytical approach, MANTRA, which allowed for different underlying allelic effects between race/ethnic groups. As the genomic regions harboring genetic signals that were shared across the different race/ethnic populations were more likely to contain functional variants, we gained power by incorporating all the samples together, at least for those loci that generalized across population groups. This approach is especially applicable to the analysis of samples of AA, since more limited LD is observed in these populations, aiding in fine-mapping. Our results may have been limited by the fact that our EA sample size was much larger than our AA sample size. Indeed, the effects of this difference in sample size require further investigation. Overall, in leveraging varying LD structures across different populations, SNPs with the strongest signals and the CSs from our trans-ethnic meta-analysis provide more precise localization of variants for future functional analysis.
MATERIALS AND METHODS
Design and samples
We conducted a meta-analysis of summary results from EA and AA GWAS. For the EA data, we used meta-analysis association results from up to 77 167 individuals in 32 cohorts, published by the GIANT consortium (5). For the AA data, we used the meta-analysis GWAS results from up to 19 744 individuals in 14 cohorts in the AAAG Consortium (6).For both EA and AA data, we obtained the results from the fixed-effects and inverse variance-weighted meta-analyses of study-specific association analyses. In this paper, we focused on the 14 loci (TBX15-WARS2, DNM3-PIGC, LYPLAL1, GRB14, NISCH-STAB1, ADAMTS9, CPEB4, LY86, VEGFA, RSPO3, NFE2L3, ITPR2-SSPN, HOXC13, ZNRF3-KREMEN1) previously identified in the participants of EA in GIANT (5).
Phenotypes
We analyzed the association of WHR, a measure of body fat distribution. For both the GIANT consortium and the AAAG Consortium, each cohort created residuals for WHR adjusted for age, age2, study site (if applicable) and BMI. The residuals were inverse normal transformed and then used as the phenotypes in association analysis within each participating cohort. Each participating cohort used principal components as needed in regression models assessing the association of a SNP to account for population stratification. Details regarding the trait creation and participating studies can be found in the original publications (5,6).
Statistical analysis
We meta-analyzed the two sets of results for the 14 previously reported loci with MANTRA software (Meta-Analysis of Trans-ethnic Association studies) (14). MANTRA is a meta-analysis approach that can be used to combine GWAS results from more than one ancestry based on the expectation of similar allelic effects between the most closely related populations (14). Technically, populations are clustered based on the average allele frequency difference by means of a Bayesian partition model and populations within the same cluster are assumed to have the same underlying allelic effect while allowing heterogeneity for populations in different clusters. The evidence in favor of association of the trait with the genetic variant is quantified with a Bayes' factor (BF) and a log10 BF of 6.1 or higher is approximately comparable to a genome-wide significance threshold of P < 5 × 10−8 (7). Specifically in our application, we conduct the fixed-effect meta-analysis within each race assuming there is no genetic heterogeneity across the samples of participating cohorts from the same race. Then, we implemented MANTRA to meta-analyze the association results from EA and AA samples.
Construction of 95% CSs
We used MANTRA results to construct a fine-mapping interval for each associated index variant (13). We constructed these intervals from analysis in the EA only set (GIANT results) and in the results from the trans-ethnic analysis. To create a 95% CS, we analyzed variants within 250 kb upstream and downstream from the variant with the index SNP. The algorithm to construct a CS is the following:
As described in MANTRA, obtain the BF value for each variant in the region.
For each SNPj, calculate the posterior probability that the SNP is driving the association signal within the region, i.e. BFj divided by summation of BF over all SNPs within the region.
Rank all SNPs within the region according to their BFs, such that BF(i) represents the ith largest BF.
Proceed down the ranked list until the accumulative posterior probability exceeds 95% of the total cumulative posterior probably for all SNPs in the locus.
Include in the 95% CS those SNPs with accumulative posterior probability of 0.95. We define the length associated with each CS as the length of the region in base pairs spanned by the SNPs retained in the specific CS.
Bioinformatics annotation
We annotated variants in the 95% CSs at the five loci that displayed a >25% decrease in 95% CS kilobase size in trans-ethnic analysis compared with the analysis in EA only: ADAMTS9, TBX15-WARS2, LYLPAL1, ITPR2-SSPN and LY86. Although the 25% decrease is an arbitrary cutoff, we chose it to focus on those CSs where there was a substantial decrease, suggesting that trans-ethnic analyses aided in fine mapping. To identify SNPs that may contribute to an association signal but were not tested in the MANTRA analysis, we also identified SNPs in strong LD with the 95% CS SNPs in either EA or AA (r2 > 0.8, AFR or EUR 1000 Genomes Phase1 version 2 release) (15). None of the interrogated variants are coding variants. The distance from each tested SNP to the nearest transcription start site of a coding gene was calculated using basic transcript annotation from GENCODE version 12 (16).
To examine variant overlap with elements from regulatory datasets, we downloaded data for selected tissue and cell types describing the locations of regions of open chromatin (DNase-seq, FAIRE-seq), histone modification signal enrichment (H3K4me1, H3K27ac, H3K4me3, H3K9ac and H3K4me2), and transcription factor binding generated by the ENCODE Consortium (17) and Roadmap Epigenomics Project (18). Data from the ENCODE Integrative Analysis were used when available (19). For consistency in processing the data across consortia, we downloaded sequence alignments from the Roadmap Epigenomics project and identified regions of enrichment using the same irreproducible discovery rate (20) pipeline as ENCODE. For tissues with only one replicate, we used only MACS2 (21) to identify regions of signal enrichment. A total of 224 datasets were collected for this analysis (Supplementary Material, Table S1).
SUPPLEMENTARY MATERIAL
FUNDING
This work is partially supported by grant NGFNPLUS 01GS0823 (T.W.W. and I.M.H.), National Institutes of Health (NIH) R01DK8925601 (I.B.B., L.A.C. and C.T.L.), R01DK072193 (K.L.M.), R21DA027040 (K.L.M.), T32GM067553 (M.L.B.) and American Heart Association 13PRE16930025 (M.L.B.).
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Andrew Morris's kindness in providing the MANTRA package and discussion.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Carey V.J., Walters E.E., Colditz G.A., Solomon C.G., Willett W.C., Rosner B.A., Speizer F.E., Manson J.E. Body fat distribution and risk of non-insulin-dependent diabetes mellitus in women. The Nurses’ Health Study. Am. J. Epidemiol. 1997;145:614–619. doi: 10.1093/oxfordjournals.aje.a009158. [DOI] [PubMed] [Google Scholar]
- 2.Lindgren C.M., Heid I.M., Randall J.C., Lamina C., Steinthorsdottir V., Qi L., Speliotes E.K., Thorleifsson G., Willer C.J., Herrera B.M., et al. Genome-wide association scan meta-analysis identifies three loci influencing adiposity and fat distribution. PLoS Genet. 2009;5:e1000508. doi: 10.1371/journal.pgen.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y., Rimm E.B., Stampfer M.J., Willett W.C., Hu F.B. Comparison of abdominal adiposity and overall obesity in predicting risk of type 2 diabetes among men. Am. J. Clin. Nutr. 2005;81:555–563. doi: 10.1093/ajcn/81.3.555. [DOI] [PubMed] [Google Scholar]
- 4.Rose K.M., Newman B., Mayer-Davis E.J., Selby J.V. Genetic and behavioral determinants of waist–hip ratio and waist circumference in women twins. Obes. Res. 1998;6:383–392. doi: 10.1002/j.1550-8528.1998.tb00369.x. [DOI] [PubMed] [Google Scholar]
- 5.Heid I.M., Jackson A.U., Randall J.C., Winkler T.W., Qi L., Steinthorsdottir V., Thorleifsson G., Zillikens M.C., Speliotes E.K., Mägi R., et al. Meta-analysis identifies 13 new loci associated with waist–hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat. Genet. 2010;42:949–960. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu C.T., Monda K.L., Taylor K.C., Lange L., Demerath E.W., Palmas W., Wojczynski M.K., Ellis J.C., Vitolins M.Z., Liu S., et al. Genome-wide association of body fat distribution in African ancestry populations suggests new loci. PLoS Genet. 2013;9:e1003681. doi: 10.1371/journal.pgen.1003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X., Chua H.X., Chen P., Ong R.T., Sim X., Zhang W., Takeuchi F., Liu X., Khor C.C., Tay W.T., et al. Comparing methods for performing trans-ethnic meta-analysis of genome-wide association studies. Hum. Mol. Genet. 2013;22:2303–2311. doi: 10.1093/hmg/ddt064. [DOI] [PubMed] [Google Scholar]
- 8.Ioannidis J.P., Ntzani E.E., Trikalinos T.A. Racial’ differences in genetic effects for complex diseases. Nat. Genet. 2004;36:1312–1318. doi: 10.1038/ng1474. [DOI] [PubMed] [Google Scholar]
- 9.Liu C.T., Garnaas M.K., Tin A., Kottgen A., Franceschini N., Peralta C.A., de Boer I.H., Lu X., Atkinson E., Ding J., et al. Genetic association for renal traits among participants of African ancestry reveals new loci for renal function. PLoS Genet. 2011;7:e1002264. doi: 10.1371/journal.pgen.1002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu C.T., Ng M.C., Rybin D., Adeyemo A., Bielinski S.J., Boerwinkle E., Borecki I., Cade B., Chen Y.D., Djousse L., et al. Transferability and fine-mapping of glucose and insulin quantitative trait loci across populations: CARe, the Candidate Gene Association Resource. Diabetologia. 2012;55:2970–2984. doi: 10.1007/s00125-012-2656-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenberg N.A., Huang L., Jewett E.M., Szpiech Z.A., Jankovic I., Boehnke M. Genome-wide association studies in diverse populations. Nat. Rev. Genet. 2010;11:356–366. doi: 10.1038/nrg2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dastani Z., Hivert M.F., Timpson N., Perry J.R., Yuan X., Scott R.A., Henneman P., Heid I.M., Kizer J.R., Lyytikäinen L.P., et al. Novel loci for adiponectin levels and their influence on type 2 diabetes and metabolic traits: a multi-ethnic meta-analysis of 45,891 individuals. PLoS Genet. 2012;8:e1002607. doi: 10.1371/journal.pgen.1002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franceschini N., van Rooij F.J., Prins B.P., Feitosa M.F., Karakas M., Eckfeldt J.H., Folsom A.R., Kopp J., Vaez A., Andrews J.S., et al. Discovery and fine mapping of serum protein loci through transethnic meta-analysis. Am. J. Hum. Genet. 2012;91:744–753. doi: 10.1016/j.ajhg.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morris A.P. Transethnic meta-analysis of genomewide association studies. Genet. Epidemiol. 2011;35:809–822. doi: 10.1002/gepi.20630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A., Consortium G.P. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S., et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome. Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Consortium E.P. A user's guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernstein B.E., Stamatoyannopoulos J.A., Costello J.F., Ren B., Milosavljevic A., Meissner A., Kellis M., Marra M.A., Beaudet A.L., Ecker J.R., et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunham I., Kundaje A., Aldred S.F., Collins P.J., Davis C.A., Doyle F., Epstein C.B., Frietze S., Harrow J., Kaul R., et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Q., Brown J.B., Huang H., Bickel P.J. Measuring reproducibility of high-throughput experiments. Ann. Appl. Stat. 2011;5:1752–1779. [Google Scholar]
- 21.Feng J., Liu T., Qin B., Zhang Y., Liu X.S. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 2012;7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.