Fig. 4.
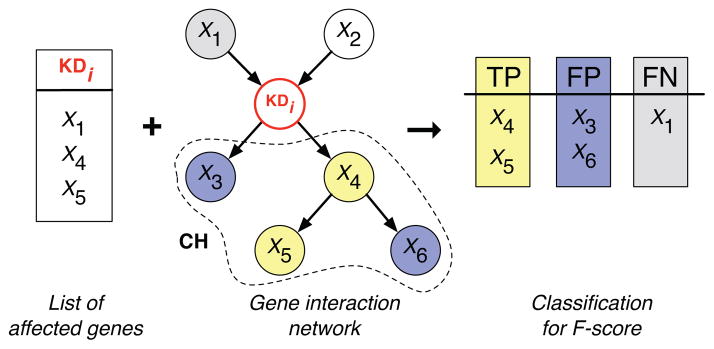
Given a set of genes affected by a knock-down gene KDi and a gene interaction network, one can define nodes as true positives (TP), false positives (FP) and false negatives (FN). In theory, all the affected genes should be inferred to be members of the knock-down’s childhood, denoted by CH. If they are found within CH, they are classified as true positives (TP). All other genes in CH are classified as false positives (FP). Affected genes that are not inferred to be in the knock-down’s CH are classified as false negatives (FN). This classification of nodes into TP, FP and FN is then used to compute a quality score, such as the F-score.
