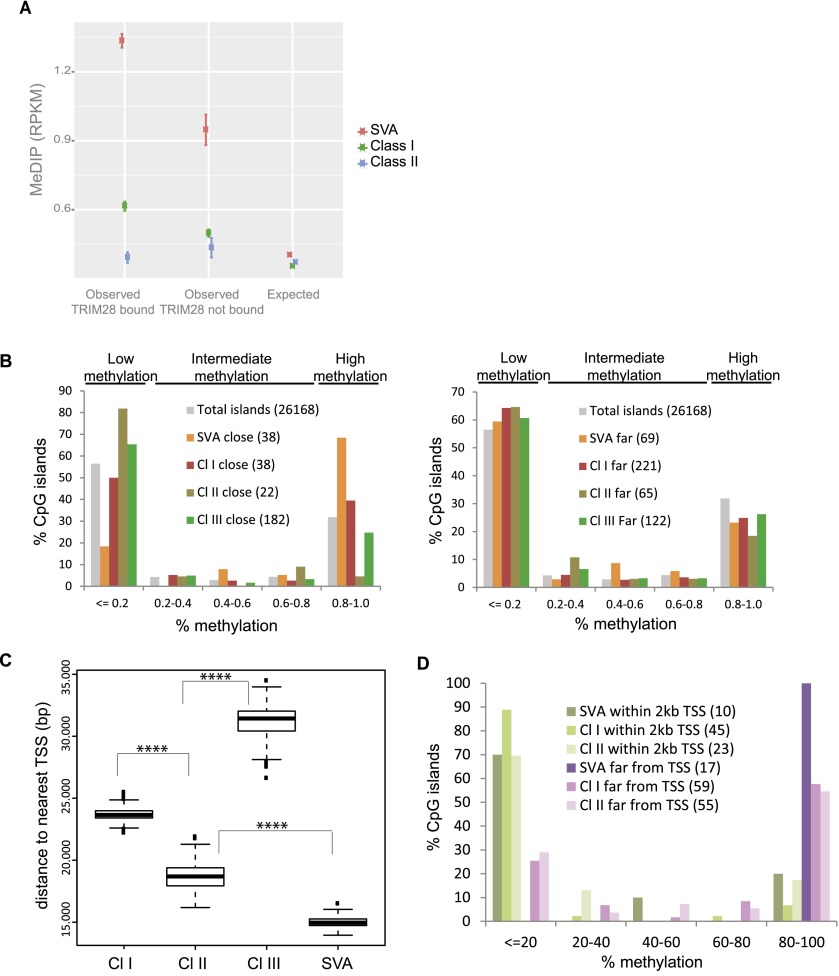
Figure 3.
TRIM28 recruitment to ERE leads to nearby CpG methylation. (A) DNA methylation on TRIM28-bound versus -unbound EREs, using publicly available MeDIP sequences. The observed mean RPKM for bound and unbound ERE is shown. For the expected values, we repositioned each ERE in each chromosome 1000 times at random. T-tests comparing bound and unbound elements gave P-values < 0.0001 for Class I and SVA EREs and P-value = 0.08 for Class II EREs. (B) Distribution of CpG islands according to methylation status and distance to indicated EREs, whether close (<2 kb for SVAs and 5 kb for HERVs to analyze similar number of CpG islands, left panel) or far (between 2 and 4 kb for SVAs, 15 and 30 kb for HERVs, right panel), using total CpG islands genome-wide as a control. In parentheses are numbers of CpG islands corresponding to each case. (C) Average distance between indicated EREs and closest TSS. P-values < 0.0001 (****) (Wilcoxon Mann-Whitney nonparametric test) are highly significant between all groups. (D) Distribution of CpG islands close to ERE, according to methylation status, whether ERE is close to (<2 kb) or far from (>20 kb) a TSS. In parentheses are numbers of CpG islands corresponding to each case.