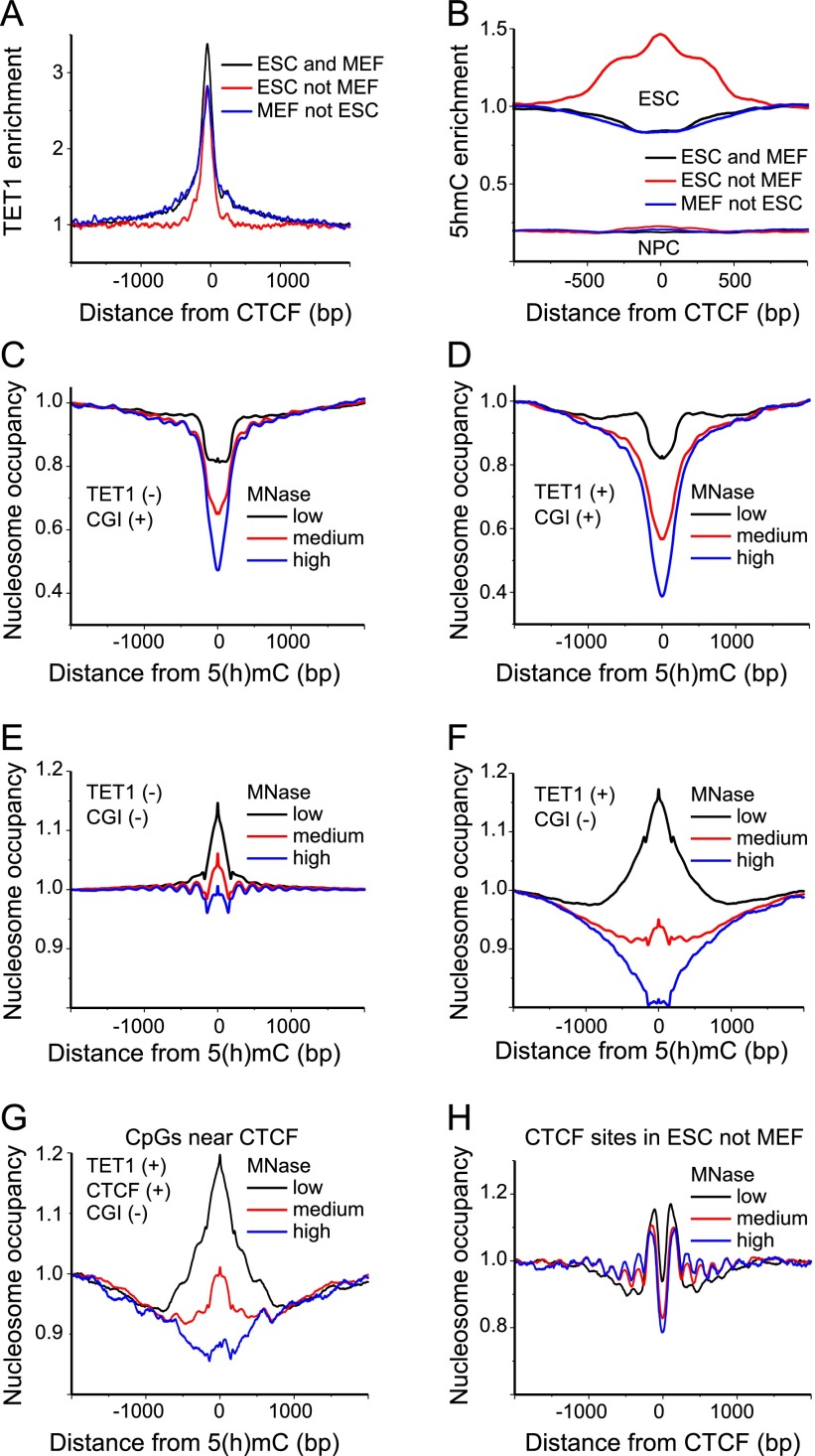
Figure 4.
TET1 binding outside CGIs is linked with labile MNase-sensitive nucleosomes. (A) Average TET1 enrichment calculated from ChIP-seq data in ESCs (Williams et al. 2011) around CTCF binding sites (Shen et al. 2012). (B) Enrichment of 5hmC calculated from hMeDIP data in ESCs and NPCs (Tan et al. 2013) around CTCF binding sites (Shen et al. 2012; Phillips-Cremins et al. 2013). (C) Average nucleosome occupancy profiles around 5(h)mC in the absence of TET1 within CGIs for low (black line), medium (red line), and high (blue line) MNase digestion in ESCs. Note that bisulfite sequencing does not distinguish between 5mC and 5hmC. (D) Same as panel C but for 5(h)mC regions enriched with TET1. (E) Nucleosome occupancy at 5(h)mC without TET1 outside of CGIs. Same color-coding as in panel C. (F) Same as panel E but for 5(h)mC regions with TET1. (G) Same as in panel F but only for (hydroxy)methylated CpGs that were within a 500 bp distance of bound CTCF. (H) Nucleosome occupancy around CTCF sites and occupied by CTCF in ESCs but not in MEFs. Same color-coding as in panel C.