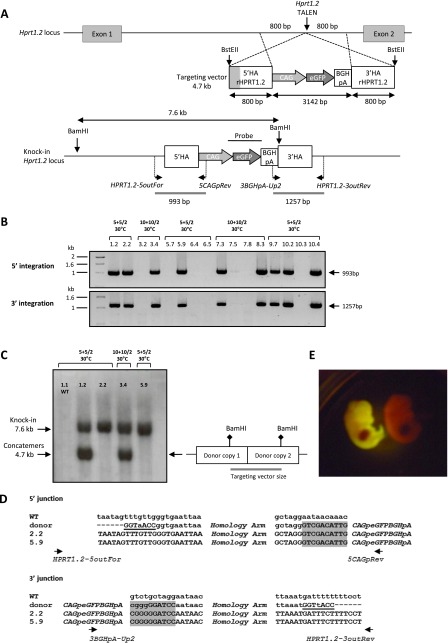
Figure 2.
Targeted integration of a GFP cassette into the Hprt1.2 locus. (A) (Upper) Diagram showing schematic representation of the rat Hprt1.2 locus, with the site of TALE nuclease action (vertical arrows), and of the targeting vector with the expression cassette CAG-eGFP-BGHpA (3142 bp) and the 5′ and 3′ homology arms (800 bp each). The homology arms are contiguous to the TALE nucleases’ cleavage point. (Gray) The sequence overlap (433 bp) between 3′ HArHPRT1.1 (cf. Supplemental Fig. S1) and 5′ HArHPRT1.2. BstEII restriction sites are indicated. (A) (Lower) Diagram showing schematic representation of the GFP cassette integration. For flanking PCR analysis, genomic DNA was PCR-amplified with primers situated for the 5′ side: upstream of the 5′ HA arm (HPRT1.2-5outFor) and in the CAG promoter (5CAGpRev); and for the 3′ side: in the BGHpA (3BGHpA-Up2) and downstream from the 3′ HA arm (HPRT1.2-3out Rev). The position of each primer and the corresponding expected size of PCR products are indicated on the schematic knock-in Hprt1.2 locus. For Southern blot analysis, genomic DNA was digested with BamHI and was probed with a GFP probe. A unique band at 7.6 kb is predicted for a correct HDR into the Hprt1.2 locus. (B) Flanking PCR analysis. Gels show the results of analyzing the 5′ and the 3′ extremities of GFP integration into the Hprt1.2 locus. A representative panel of 16 animals is illustrated, showing the expected bands of 993 bp using the 5′ pair of primers (HPRT1.2-5outFor + 5CAGpRev) and of 1257 bp using the 3′ pair of primers (3BGHpA-Up2 + HPRT1.2-3outRev). The microinjection conditions in terms of mRNA and DNA concentrations, as well as embryo incubation temperatures, are above each animal. (C) Southern blot analysis following BamH1 DNA digestion for the analysis of transgene site-specific integration into the Hprt1.2 locus using a GFP probe. Four Hprt1.2-GFP+ rats with positive junction PCRs showing a unique band at 7.6 kb for a HDR with a single copy of the transgene, or 7.6 kb and 4.7 kb (the size of the transgene) bands when HDR involved transgene concatemers. The diagram at the right explains the expected size of the concatemers once linearized. The absence of additional bands demonstrates that there are no RI integration events in these animals. As a negative control, an offspring rat with no GFP expression and negative for GFP PCR (1.1) showed no bands. (D) Sequence comparison at the 5′ and 3′ junctions, with wild-type genomic DNA and donor DNA sequences, of two representative embryos (2.2 and 5.9). In donor DNA, the presence of the BstEII site is indicated in 5′ and 3′ (underlined). The 5′ and 3′ ends of the expression cassette are colored in gray. (E) Representative E15 Hprt1.2 HDR-GFP embryos using a Dark Reader Spot Lamp.