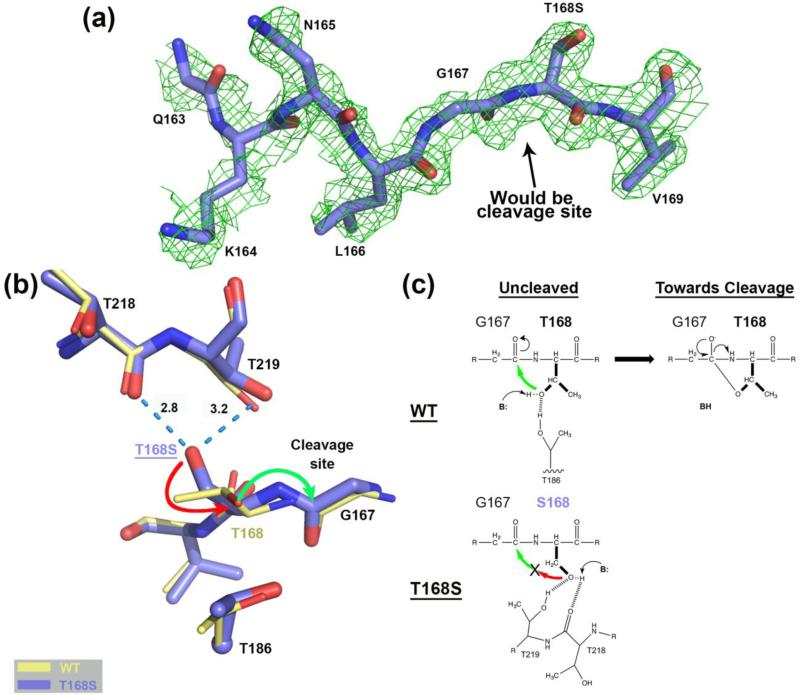
FIGURE 2.
Comparison of the hASNase3 T168S mutant to the WT uncleaved high-resolution structure. (a) Simulated annealing omit map (2Fo-Fc) from CNS contoured at 1σ around residues Gln163 to Val169 of the T168S structure Protomer A. (b) WT hASNase3 uncleaved structure (thin yellow sticks) and the T168S mutant (thick blue sticks) are superimposed and a close-up of the cleavage site is presented. Serine at position 168 interacts with the carbonyl of Thr218 and the side chain of Thr219 (dashed blue lines with distances in Angstroms). The side chain of Ser168 would have to rotate 180° (red arrow) to be able to proceed to nucleophilic attack on Gly167 carbonyl (green arrow) and break the peptide bond. Thr186 side chain is also represented with its α-carbon displayed as a sphere. (c) On top, proposed scheme showing the WT hASNase3 nucleophilic attack from Thr168 side chain on the Gly167 carbonyl with B: representing the base responsible for additional Thr168 activation. At the bottom, the same reaction within the T168S mutant is hampered by Ser168 side chain being maintained by interactions with threonines 218 and 219 and thus oriented in a catalytically non-competent conformation.