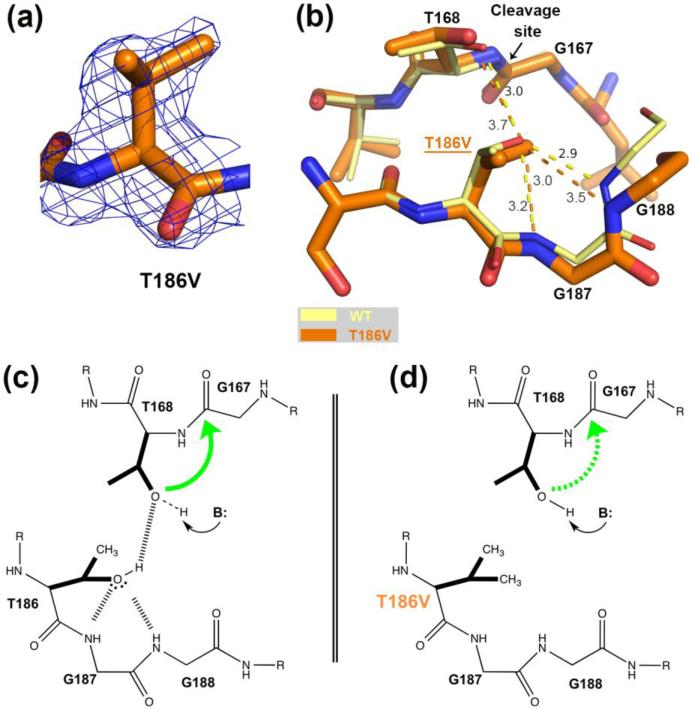
FIGURE 3.
Comparison of the hASNase3 T186V mutant to the WT uncleaved high-resolution structure. (a) Fo-Fc omit map contoured at 2.2σ around valine at position 186. Residue 186 from Protomer A was removed from the model that then underwent several rounds of refinement to eliminate model bias. (b) WT hASNase3 uncleaved structure (thin yellow sticks) and the T186V mutant (thick orange sticks) are superimposed and a close-up of the cleavage site is presented. In the WT enzyme, the hydroxyl group of Thr186 hydroxyl is oriented by the main chain NH atoms of highly conserved glycines 187 and 188 (3.0 Å and 2.9 Å respectively) and by Thr168 (3.0 Å). Dashed lines represent hydrogen bonding interactions with distances given in Angstroms. When the isosteric valine replaces the threonine at position 186, the side chain maintains the same orientation but the distance to Thr168 increases to 3.7 Å in order to avoid steric clashes from their respective side chains. (c) Scheme proposing a cleavage mechanism for WT hASNase3 in which Thr186 builds a hydrogen bond donor interaction to the Thr168 hydroxyl and hydrogen bond acceptor interactions to the Gly187/188 NH atoms. As a result of accepting a proton from Thr186, the Thr168 hydroxyl becomes more likely to be activated by a base (B:). The green arrow represents the nucleophilic attack from Thr168 hydroxyl on Gly167 carbonyl. (d) In the T186V mutant, valine at position 186 is unable to participate in hydrogen bond interactions. As a result, Thr168 activation is more difficult to attain (dashed green arrow).