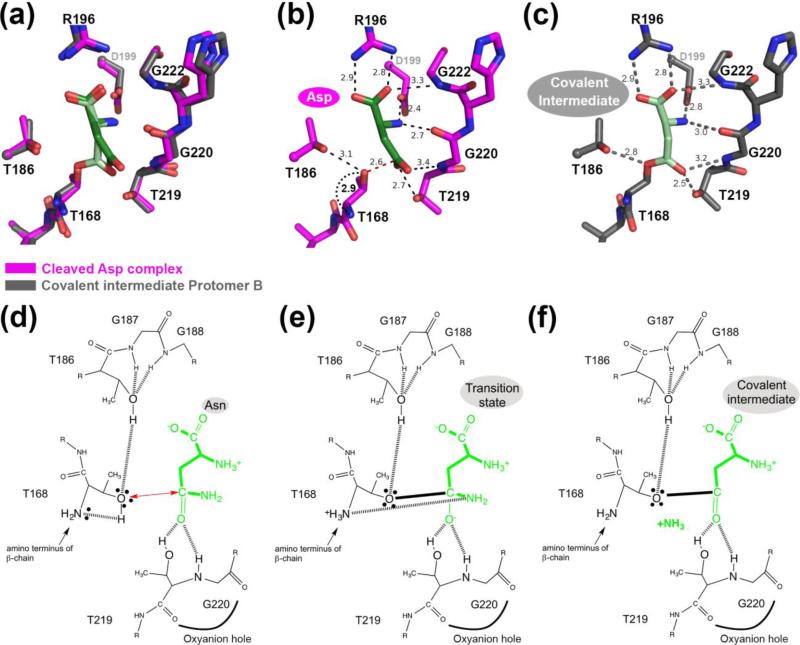
FIGURE 6.
Comparison of fully cleaved WT hASNase3 in complex with Asp and in the covalent intermediate state. (a) Fully cleaved WT hASNase3 structure in complex with Asp (PDB ID 4GDW; enzyme in magenta, Asp in dark green) and the WT covalent intermediate hASNase3 protomer B structure (enzyme in grey and covalently linked substrate in lighter green) are superimposed and a closeup of the active site is presented. (b) WT hASNase3 active site from the fully cleaved Asp complex structure. Black dashed lines indicate interactions between Asp (dark green) and hASNase3. The red dashed line shows the distance between the Thr168 hydroxyl group and the Asp β-carboxylate carbon atom (2.6 Å). The curved dotted black line indicates the distance between the freed amino group from Thr168 activating its hydroxyl for nucleophilic attack on the substrate side chain (all distances are in Angstroms). (c) WT hASNase3 active site from the covalent intermediate protomer B structure. Grey dashed lines indicate interactions between the covalently linked substrate (light green) and hASNase3. (d) hASNase3 asparaginase mechanism. After enzyme cleavage and binding of Asn, the freed amino group of Thr168 activates its side chain, allowing for nucleophilic attack on the substrate side chain (red arrow). This reaction is facilitated by the interaction between Thr186 and Thr168 side chains. (e) The resulting negatively charged tetrahedral intermediate is stabilized by the oxyanion hole composed of Thr219 and Gly220 in the transition state. (f) After release of ammonia, the being-hydrolyzed amino acid remains covalently linked to Thr168 in a state we refer to as the covalent intermediate.