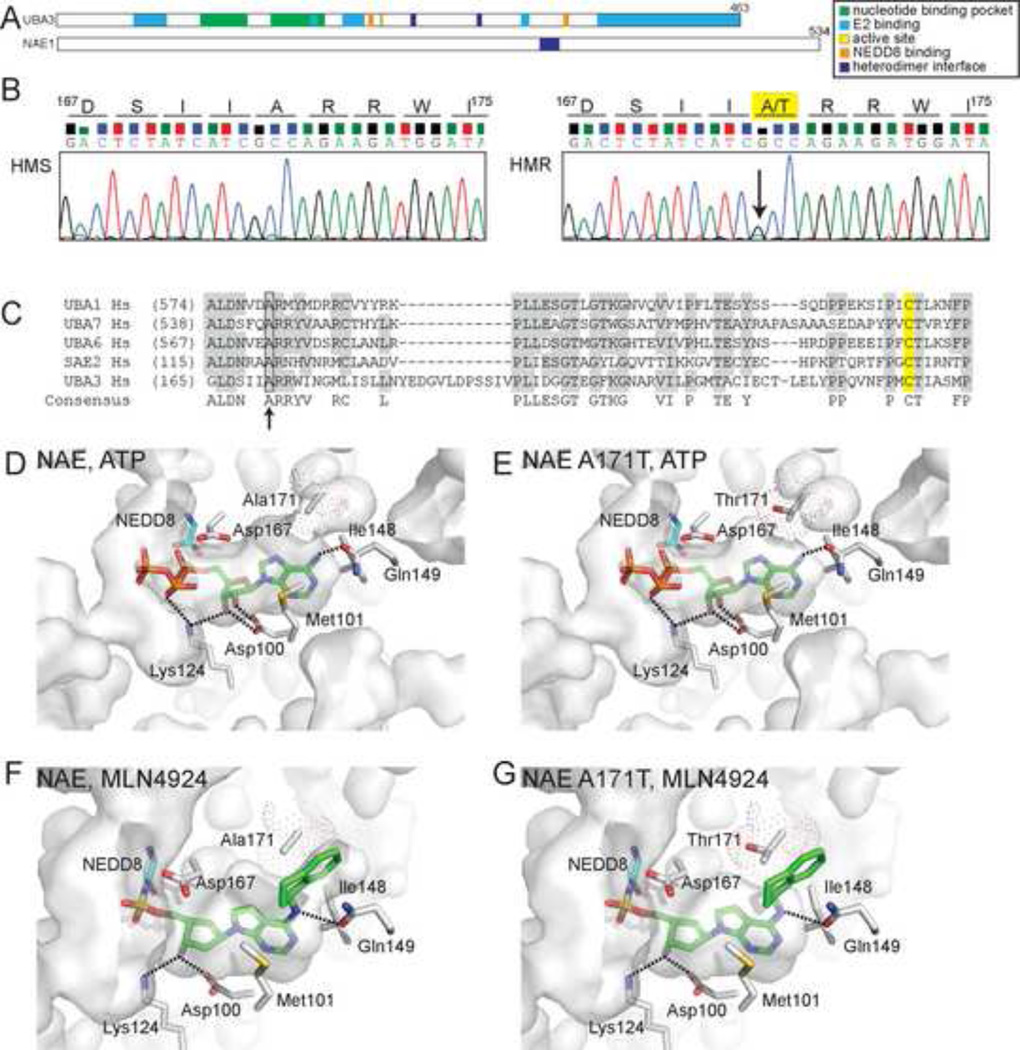
Figure 2. HMR cells are heterozygous for a single nucleotide transition within the coding sequence of the UBA3 nucleotide binding pocket.
(A) Domain structure of NAE subunits, UBA3 (UBE1C) and NAE1 (ULA1, APPBP1). (B) Chromatograms from sequencing reactions analyzing genomic DNA from HMS and HMR cells determined that HMR cells are heterozygous for a G to A transition (indicated by arrow) that alters an amino acid within the UBA3 nucleotide binding pocket. This transition results in the conversion of alanine 171 to threonine (A171T) in the UBA3 open reading frame. See Figure S2 for sequences of cDNAs and extended genomic DNA sequence. (C) A sequence alignment of canonical E1s indicates similarly positioned alanine residues (indicated by arrow) found in other E1s. The catalytic cysteines are highlighted in yellow. Molecular models of the NAE nucleotide binding pocket with ATP bound to UBA3 (D) or UBA3 A171T (E) and MLN4924-NEDD8 bound to UBA3 (F) or UBA3 A171T (G). The solvent accessible nucleotide binding pocket is indicated as are side chains important for MLN4924 binding. Dot surface representation for A171 and computer modeled A171T is shown with only the rotamer with the highest frequency of occurrence in proteins. Computer modeled A171T rotamers in (E) had significant van der Waals overlaps with other residues in UBA3, suggesting it could alter the nucleotide binding pocket. Interestingly, the two major A171T rotamers modeled in (G) had significant van der Waals overlap with the aminoindane of MLN4924, but only minor contacts with other UBA3 residues. These models are based on Protein Data Bank accession codes 1R4N (D, E) and 3GZN (F, G).