Figure 4.
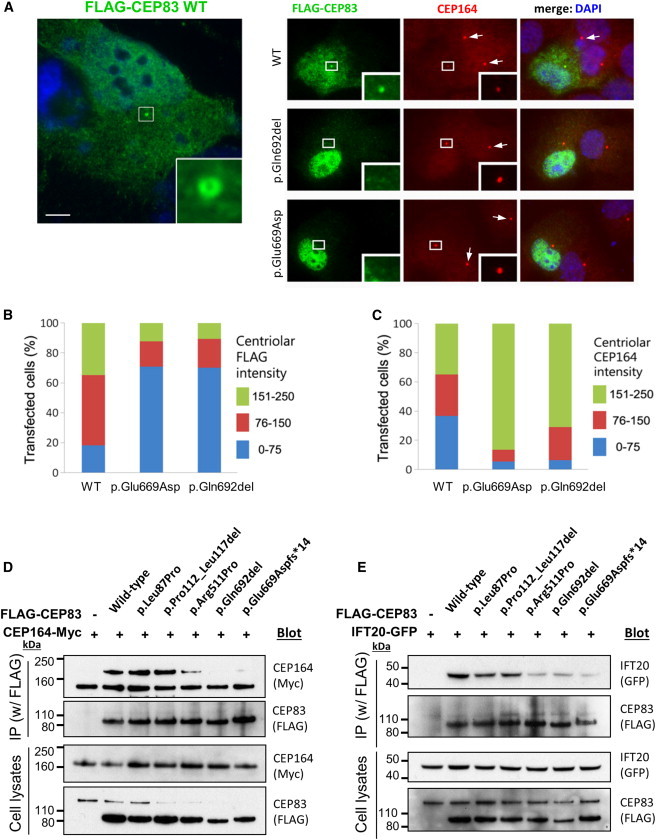
CEP83 Mutations Affect Centrosomal Localization and Interaction of CEP83 with CEP164
(A) RPE1 cells were transiently transfected (Lipofectamine LTX, Life Technologies) with plasmids encoding for FLAG-tagged wild-type (WT) or c.2007del (p.Glu669Aspfs∗14) or c.2075_2077del (p.Gln692del) CEP83 variants, fixed, and stained for FLAG (green; mouse monoclonal [Sigma, F3165]) and CEP164 (red). Scale bar represents 5 μm.
(B and C) Levels of CEP164 (C) and FLAG (CEP83, B) staining at the centrosome were quantified with ImageJ. Centriolar CEP164 intensity is decreased in cells transfected with CEP83 WT compared to cells transfected with CEP83 p.Glu669Aspfs∗14 and p.Gln692del variants (arrows).
(D and E) NIH 3T3 cells were transiently cotransfected with plasmids encoding for FLAG-tagged WT and variant forms of CEP83, Cep164-myc, and IFT20-GFP. Cell lysates were immunoprecipitated with an anti-FLAG antibody (mouse monoclonal, Sigma, M2). Cell lysates (bottom) and immunoprecipitates (IP, top) were analyzed by immunoblotting with antibodies against myc (CEP164, rabbit polyclonal [Santa Cruz Biotechnology, C-19]), FLAG (CEP83, rabbit polyclonal [Cell Signaling Technology, #2368]), and GFP (IFT20, rabbit polyclonal [Life Technologies, A-6455]) as indicated. Note that three C-terminal CEP83 variants affected the interaction with CEP164 and IFT120, whereas two N-terminal CEP83 variants did not.
