Figure 3.
Structural Modeling of MAB21L2 and Prediction of Nucleotidyltransferase Activity
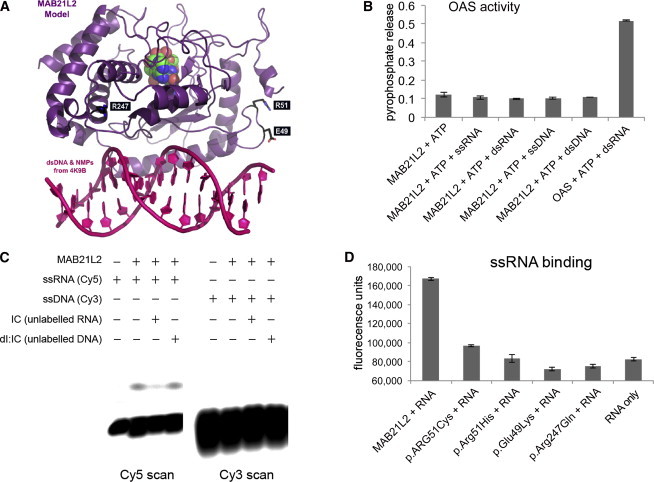
(A) A model of MAB21L2 was generated with PDB 4K9B as a template and is shown in purple; the nucleotide monophosphates are shown in green, blue, and red. This analysis suggests that MAB21L2 has both a nucleotidyltransferase active site and a DNA- and/or RNA-binding domain (double-stranded DNA is shown in pink in the foreground). The position of the residues that were altered in the affected individuals is shown in white text in black boxes. The arginine residues (Arg51 [R51] and Arg247 [R247]) are highlighted in blue, and the glutamic acid residue (Glu49 [E49]) is shown in orange.
(B) A graph showing the absence of OAS-like activity in purified MAB21L2. When OAS protein purified in the same way as MAB21L2 was incubated with ATP and double-stranded RNA (dsRNA), significant pyrophosphate release was detected, indicating nucleotidyltransferase activity. MAB21L2 showed no activity above background with ATP (or other nucleoside triphosphates [Figure S2]) using dsRNA, double-stranded DNA, ssRNA, or ssDNA as an activator.
(C) An electromobility shift assay (EMSA) using fluorescently labeled I:C oligonucleotides shows binding of wild-type MAB21L2 to ssRNA, but not ssDNA. The ssRNA binding could be completed efficiently with unlabeled ssRNA, but not ssDNA.
(D) Solution-based assay showing that wild-type MAB21L2 could efficiently bind a digoxigenin-labeled ssRNA molecule (this was an antisense riboprobe against FZD5, but all probes tested behaved in an identical fashion). None of the altered proteins could bind the ssRNA probe at levels above background.
The error bars in (B) and (D) represent SE. Each experiment represents readings from two biological replicates, and all experiments were repeated twice.
