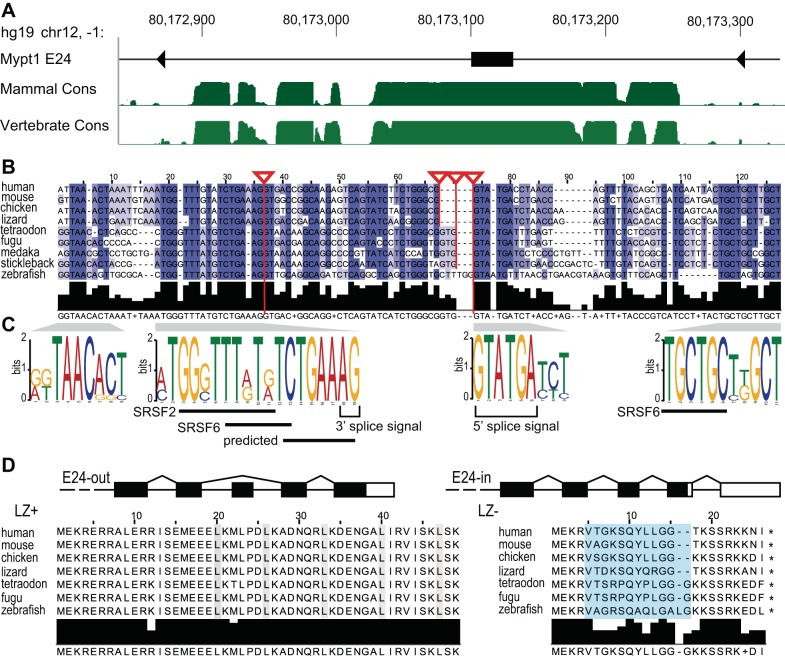
Fig. 2.
Phylogenetic conservation of isoforms of Mypt1 generated by alternative splicing of Exon 24. A: conservation of the myosin targeting subunit 1 (Mypt1) alternative exon E24 and the flanking intronic region is shown by the phastCons track on the UCSC Genome Browser in the human hg19/GRCh37 genome release. The phastCons track is a multiple alignment of 46 vertebrate species (“Vertebrate Cons”) and a subset of 33 placental mammal (“Mammal Cons”) that estimates the probability of individual nucleotides belonging to a conserved element, by considering both the individual alignment column and its flanking columns. The higher the green bars the more likely the region belongs to a conserved element. B: alignment and coloring based on percent identity of Mypt1 E24 in human, mouse, chicken, and lizard, and the predicted alternative exon in fish demonstrates regions of high conservation immediately flanking the exon. Red lines attached to triangles highlight known and predicted (in fish) splice sites. Conserved sequences were identified and analyzed for splicing cis-regulatory elements as described in methods. C: exon-intron structure of the 3′ end of the Mypt1 gene is shown. Alternative splicing of the 31 nt E24 changes the reading frame. Amino acid sequence alignment of the Mypt1 COOH-terminus for the E24-out/leucine zipper (LZ)+ and the E24-in/LZ− isoforms, demonstrates phylogenetic conservation, with LZ+ more conserved than LZ−. The leucines of the LZ motif for the E24-out isoform (left) are highlighted in gray. The amino acids that are coded by the alternative exon E24 are highlighted in blue.