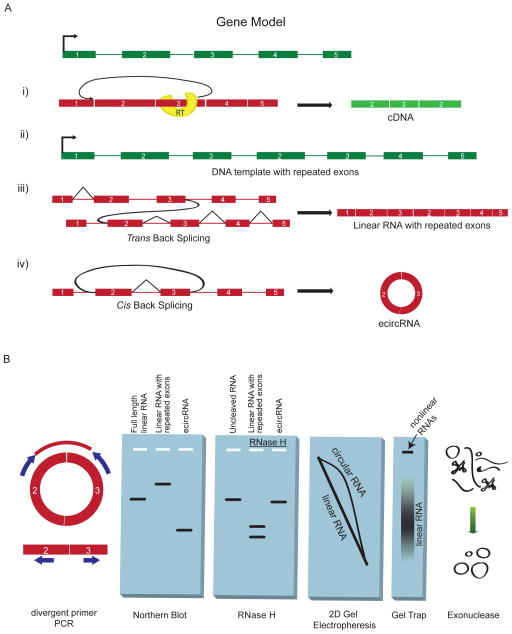
Figure 1. Splicing products and methods for detection.
(A) Several mechanisms can form an apparent backsplice, illustrated here for a gene model in which exons are shown as rectangles, introns as thin lines and the transcription start site as a right-angled arrow. DNA is in green and RNA is in red. (i) Reverse transcriptase (RT) template switching, in which the RT enzyme transcribes another copy of an upstream exon. (ii) Tandem duplications in the DNA template resulting in repeated exons. (iii) Trans-backsplicing, in which one RNA molecule is spliced to another (shown by curved black line). Regular splicing events are shown as angled black lines. (iv) Exonic circRNAs can form by cis-backsplicing in which exons from the same RNA molecule are spliced together to form a circle (backsplicing shown by curved black line). (B) Molecular assays to distinguish exonic circRNA from other backsplice products, and diagrammatic representation of the expected results. (i) Divergent primers that would amplify in outward facing directions with respect to genomic sequence become properly inward facing an produce discreet amplicons when a backsplice connects outside sequences. (ii) The expected migration distance of a canonical linear RNA in a denaturing agarose gel, as well as the relative migration of exonic circRNA and RNAs resulting from trans-splicing or tandem duplication. (iii) Migration of RNA through an agarose gel before and after RNAse H treatment. Circular RNA, uniquely, results in a single band after being cut once. (iv) Two dimensional gel electrophoresis through two differently crosslinked polyacrylamide gels separates circular RNAs into an off-diagonal curve. (v) Gel-trapping holds circular RNAs in the well of an electrophoresis gel as linear RNAs migrate away. (vi) Exonuclease enrichment degrades linear RNAs while leaving a pool enriched for circular RNA.