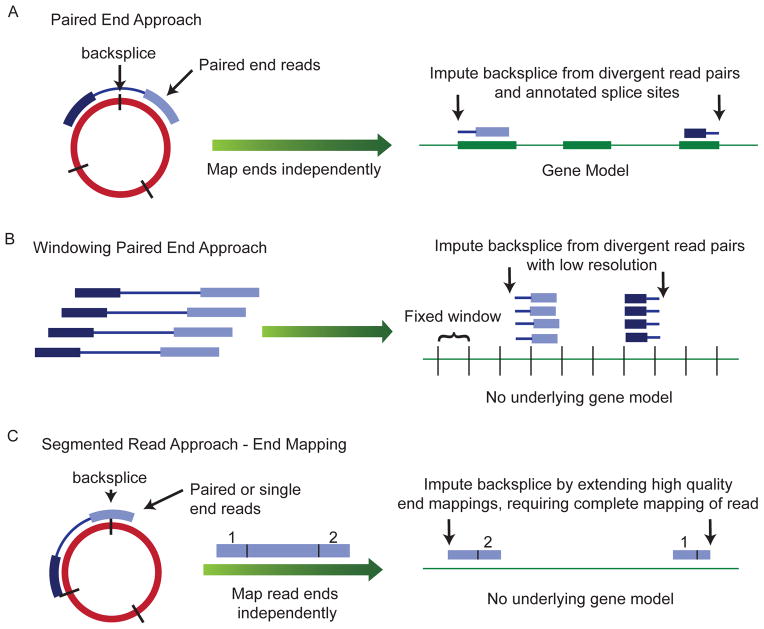
Figure 2. Sequencing-based methods for identification of exonic circRNA.
Several informatics methods have been used to identify the locations of backsplices using deep sequencing data from rRNA depleted RNA. (A) Paired-end reads may be mapped separately to the annotated transcriptome and the location of a backsplice inferred when the pairs have opposite orientations on either side of one or more splice sites. (B) Paired-end reads may be mapped directly to the genome and multiple reads that map out of genomic order suggest there might be a nearby backsplice. Stronger evidence is provided by the presence of multiple such reads accumulating in fixed “windows” of set length tiled across the genome. (C) A read can be segmented so that different parts of the read can be mapped to different parts of the genome; this allows backsplices to be mapped at nucleotide resolution without existing annotations.