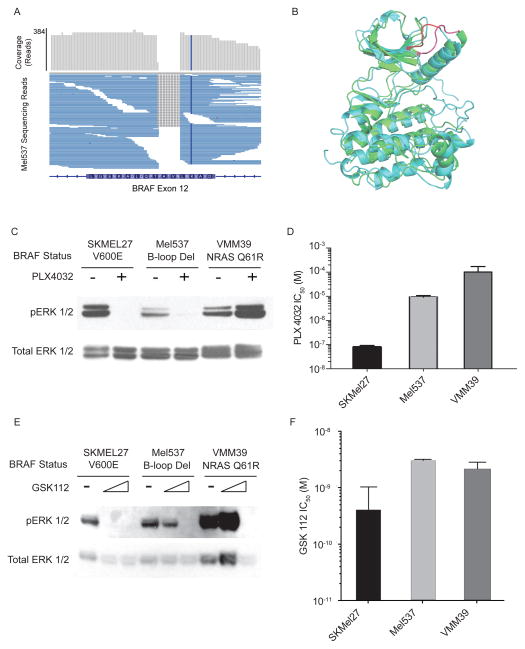
Figure 2. Non-Frameshift BRAF Mutation Sensitive to PLX4032.
(A) Mel537 cell line aggregate read depth and mapped reads show homozygous non-frameshift deletion mutation in exon 12 of BRAF. (B) Structural alignment of inhibitor bound BRAF structure (CYAN) to inhibitor bound EGFR (GREEN) showing structural homology of the BRAF exon 12 deletion (RED) and know activating EGFR exon 19 deletions (PINK). (C) ERK and activated ERK (pERK) blots in serum-starved lines with and without PLX4032 treatment. (D) PLX4032 sensitivity of SKMEL27 (BRAF V600E), Mel537, and VMM39 (NRAS Q61R). (E) ERK and activated ERK (pERK) blots in serum-starved lines with and without MEK inhibition by GSK112. (F) GSK112 sensitivity of SKMEL27, Mel537 and VMM39.