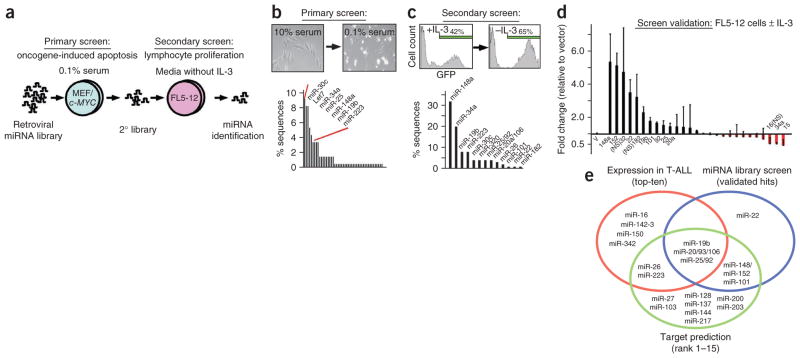
Figure 2.
Pooled library screen for oncogenic miRNAs. (a) Schematic of the screening protocol: the primary screen selected for bypass of oncogene-induced apoptosis and the secondary screen selected for lymphocyte proliferation in the absence of IL-3. (b) Primary screen: micrograph illustrating c-MYC–induced apoptosis in MEFs (inset). Shown below is the percentage of miRNA sequences retrieved from surviving and adherent cells. Briefly, we isolated DNA and amplified the integrated miRNA(s) by PCR, sub-cloned it, picked ~100 clones picked and sequenced the integrated miRNA. (c) Secondary screen: enrichment of FL5-12 cells expressing the secondary miRNA library and GFP upon IL-3 depletion (inset). Shown below is the percentage of miRNA sequences retrieved from FL5-12 cells after IL-3 depletion. (d) Screen validation: shown is the mean fold change and the s.d. of miRNA- or GFP-expressing FL5-12 cells before and after IL-3 depletion (results of three independent experiments). (e) Summary of interim results: the ten most highly expressed miRNAs in human T-ALL (red circle), the validated ‘hits’ in the miRNA screen (blue circle) and miRNAs that bind tumor suppressor genes implicated in T-ALL (green circle).