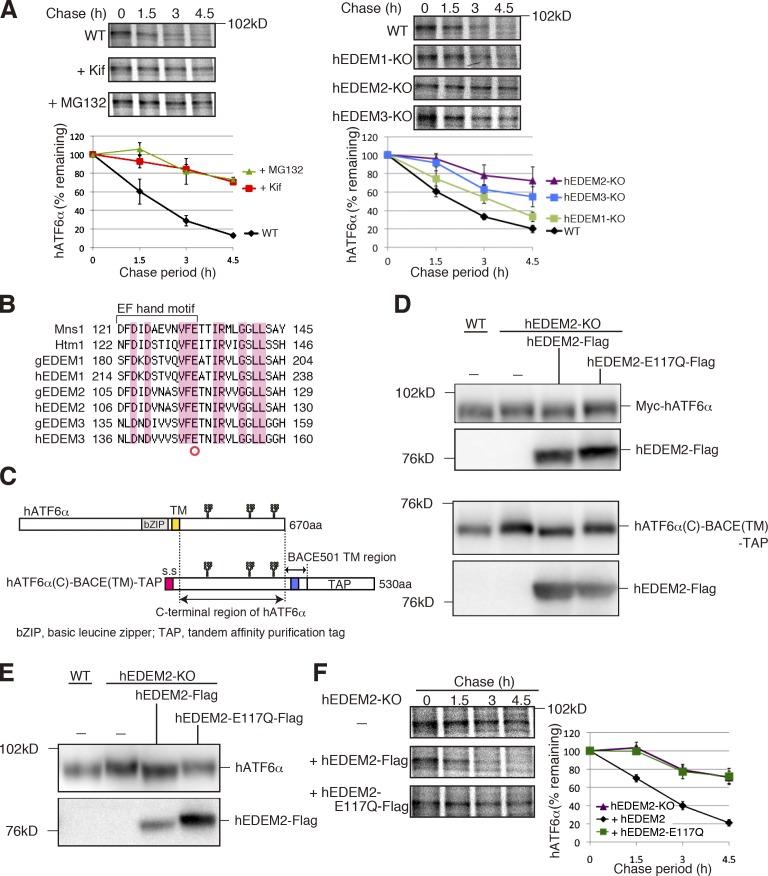
Figure 4.
Requirement for α1,2-mannosidase activity of hEDEM2 in human gpERAD. (A) Pulse chase to determine the degradation rate of endogenous hATF6α in WT HCT116 cells with or without kifunensine or MG132 treatment and in three hEDEM-KO cells (n = 3). (B) Alignment of amino acid sequences around the EF hand motif, which is essential for the α1,2-mannosidase activity of Mns1, with those of Htm1 and their homologues in chicken and human. Identical amino acids are highlighted. (C) Schematic structure of hATF6α and hATF6α(C)-BACE(TM)-TAP. Potential N-glycosylation sites are shown schematically. (D) Immunoblotting of cell lysates prepared from WT and hEDEM2-KO HCT116 cells in which Myc-hATF6α (top) or hATF6α(C)-BACE(TM)-TAP (bottom) was expressed by transfection together with hEDEM2-Flag or hEDEM2-E117Q-Flag, using anti–c-myc and anti-Flag antibodies. (E) Immunoblottig of cell lysates prepared from WT HCT116, hEDEM2-KO, and hEDEM2-KO cells stably expressing hEDEM2-Flag or hEDEM2-E117Q-Flag using anti-human ATF6α and anti-Flag antibodies. (F) Pulse chase to determine the degradation rate of endogenous hATF6α in HCT116 hEDEM2-KO and hEDEM2-KO cells stably expressing hEDEM2-Flag or hEDEM2-E117Q-Flag (n = 3).