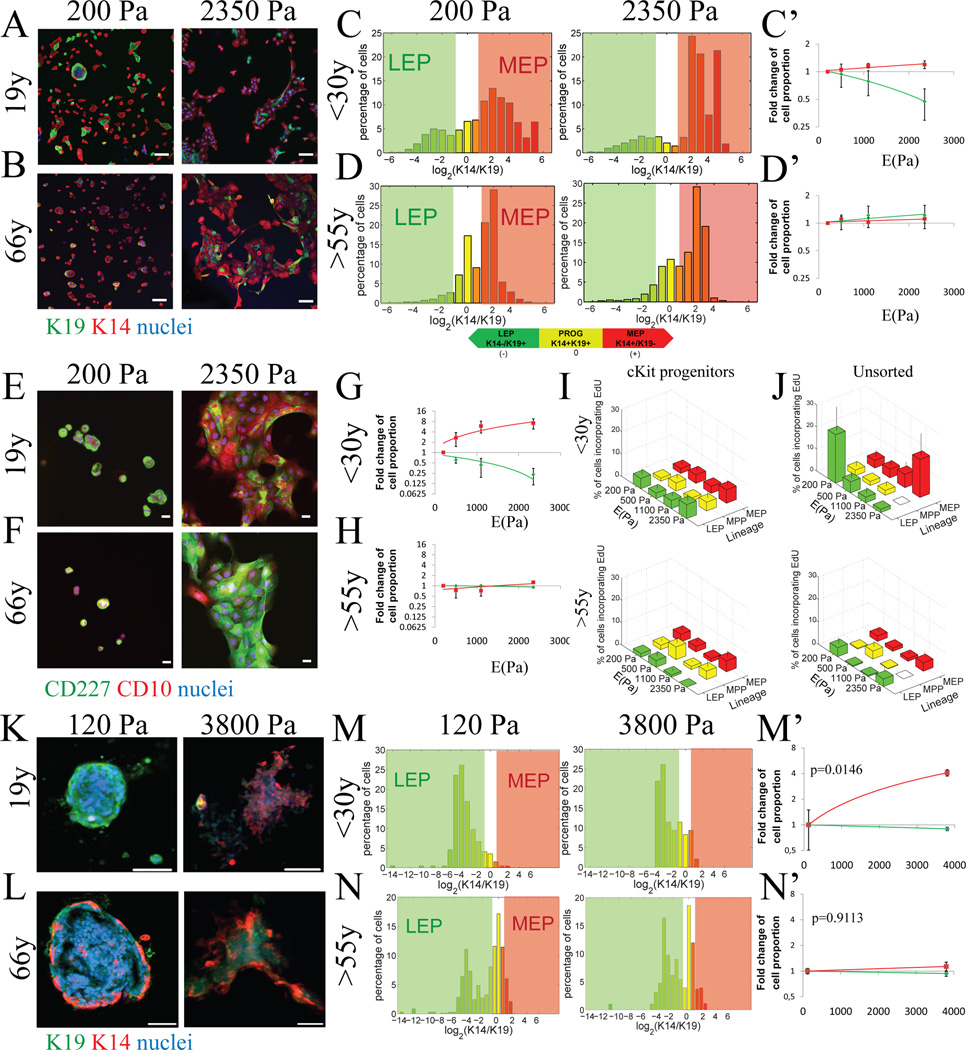
Figure 1. Aging alters differentiation patterns in response to substrate elastic modulus.
Representative IF analysis of progenitors from young (240L, 19y) and (B) an older (122L, 66y) strains stained for K19 (green), K14 (red) and DAPI (blue) after 48h of culture on PA gels with increasing elastic modulus (E(Pa)). Bars represent 100µm. (C & D) Histograms represent log2-transformed ratios of K14 to K19 protein expression in single cells on 2D PA gels with increasing stiffness, histograms are heat mapped to indicate cells with the phenotypes of K14-/K19+ LEPs (green), K14+/K19+ progenitors (yellow), and K14+/K19-MEPs (red). Corresponding linear regression plots of LEP and MEP proportions as a function of modulus are shown for (C) women <30y (LEP p=0.0429, r2= 0.2086, MEP p=0.0475, r2= 0.2009 n=5) and (D’) women >55 (LEP p=0.4812, r2= 0.0296, MEP p=0.5138, r2= 0.0240, n=5). Regressions are fold change of lineage proportions compared to cKit+ on 200Pa condition ± s.e.m. (E) Representative IF analysis of progenitors from young (240L, 19y) and (F) an older (122L, 66y) strains stained for CD227 (green), CD10 (red) and DAPI (blue) after 48h of culture on PA gels with increasing elastic modulus. Bars represent 20µm. (G) Linear regression of fold change of LEP (CD227+/CD10-) and MEP (CD227-/CD10+) proportions as function of elastic modulus in women <30y (LEP p=0.0080, r2= 0.5219, MEP p=0.0198, r2= 0.4340 n=3) and (H) women >55y (LEP p=0.4689, r2= 0.0536, MEP p=0.1937, r2= 0.5219, n=3). Linear regressions are of fold change of lineage proportions normal to 200Pa condition ± s.e.m. Percentage of cells incorporating EdU as function of lineages and stiffness in (I) cKit+ HMEC and (J) unsorted HMEC (MEP from women <30y linear regression p=0.0270, r2=0.9467, LEP from women <30y t-test 200Pa vs 2350Pa p=0.0153, n=5). Data are means ± s.e.m. (K and L) Representative immunofluorescence of K19 (green), K14 (red) and DAPI (blue) of passage 1 cKit+ HMEC from a 19y and a 65y woman after 7 days of culture encapsulated in 3D hyaluronic acid (HA) gels. Bars represent 20nm. (M and N) Histograms represent log2-transformed ratios of K14 to K19 protein expression in single cells in 3D HA gels, histograms are heat mapped to indicate cells with the phenotypes of K14-/K19+ LEPs (green), K14+/K19+ progenitors (yellow), and K14+/K19- MEPs (red). (M’ and N’) Fold changes of lineage proportions normal to 120Pa condition (Chi-squared test women <30y p=0.0146, women >55y p=0.9113). See also Figure S2, S3, S4 and Table S2.