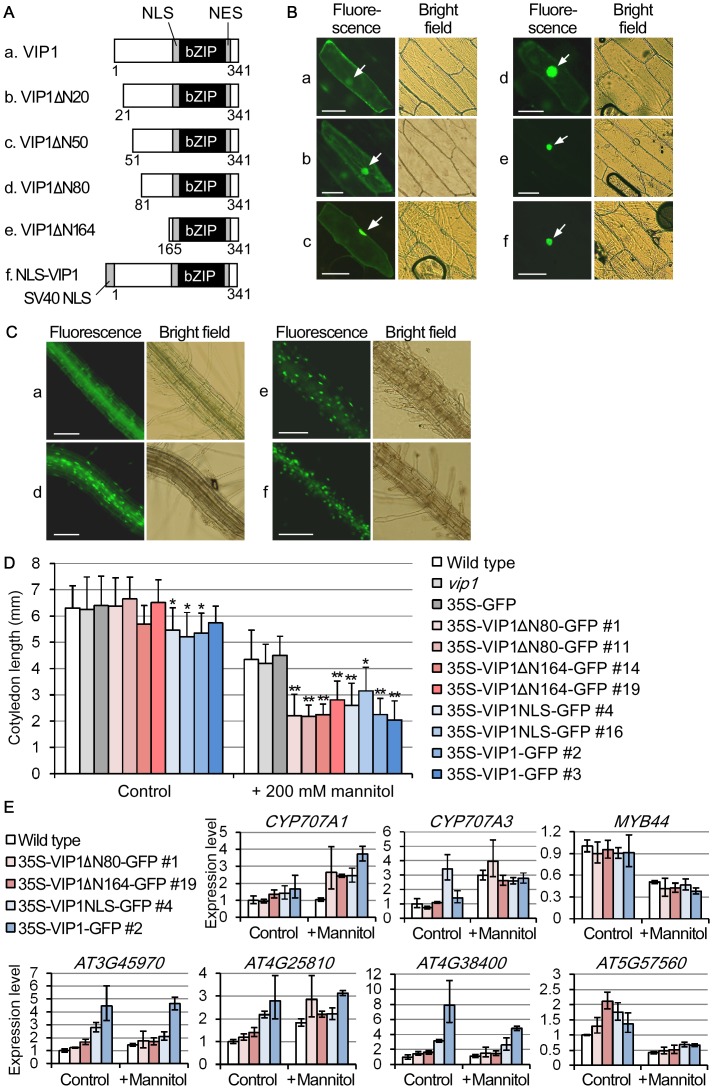
Figure 3. Generation and characterization of constitutively nuclear-localized variants of VIP1.
(A) Schematic representations of the VIP1 variants. Numbers indicate amino acid positions of full-length VIP1. Lowercase letters (a–f) correspond to those in the panels B–D. NLS: nuclear localization signal; NES: nuclear export signal. (B) Subcellular localization of the VIP1 variants in onion cells. The indicated forms of VIP1 (a–f) were expressed as GFP-fused proteins in onion epidermal cells. For each construct, more than 10 cells were observed, and a representative image is shown. Scale bars = 100 µm. (C) Subcellular localization of the VIP1 variants in Arabidopsis. Roots of the transgenic Arabidopsis plants expressing GFP-fused VIP1 variants (forms a, d, e and f) were observed without being treated by a hypotonic solution. For each genotype, more than 10 plants were used for observation, and a representative image is shown. Scale bars = 100 µm. (D) Expression of the VIP1 variants retards the cotyledon growth under a mannitol-stressed condition. The transgenic plants expressing the GFP-fused VIP1 variants (35S-VIP1ΔN80-GFP, for example) were grown in the presence of 0 (Control) or 200 mM mannitol for 10 days, and their cotyledon lengths were measured. vip1, which lacks the 3′ region of the VIP1 transcript because of a T-DNA insertion in the genomic region of VIP1 [13], is shown as control. Values are means ± SD. (n = 15–20). *: P<0.05; **: P<0.01 vs. the wild type in Student's t-test. (E) Quantitative RT-PCR analyses of expression of the genes with promoters containing AGCTGT/G in the transgenic plants expressing the VIP1 variants. The transgenic plants were grown in the presence of 0 (Control) or 200 mM mannitol (+ Mannitol) for 18 days, and sampled for RNA extraction and cDNA synthesis. Relative expression levels were calculated by the comparative cycle threshold (CT) method using UBQ5 as an internal control gene. Data are means of three biological replicates. Error bars indicate SD.