Figure 4. SMYD3 methylation of MAP3K2 activates MAP Kinase signaling pathways and repels PP2A.
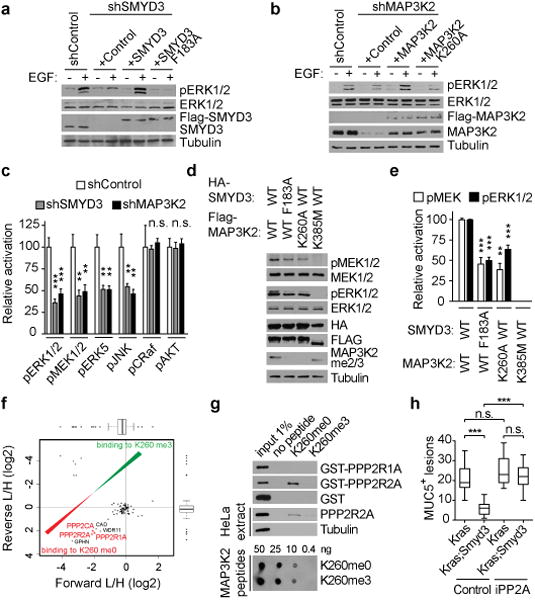
a, SMYD3 catalytic activity is required for ERK1/2 activation in LAC cells. Immunoblots with the indicated antibodies of LKR10 cells lysates depleted for SMYD3 and reconstituted with either active or inactive Flag-SMYD3 as indicated. Stimulation: EGF treatment for 15 min at 25 ng/μl. b, MAP3K2 methylation is required for ERK1/2 activation in LAC cells. Immunoblot with the indicated antibodies of LKR10 cells lysates treated as in (a) depleted for MAP3K2 and reconstituted with either wild-type or K260A mutant Flag-MAP3K2. c, SMYD3 and MAP3K2 regulate multiple overlapping MAP kinase pathway proteins. Quantitation of the indicated activated kinase signals in shSMYD3 and shMAP3K2 relative to shControl cell lysates based on three independent biological replica treated as in Extended Data Fig. 7a. d, SMYD3 catalytic activity promotes MAP3K2-induced phosphorylation of MEK1/2. Immunoblots with the indicated antibodies of 293T cells lysates transfected with Flag-MAP3K2 and HA-SMYD3 wild-type and derivatives as indicated. MAP3K2 K385 is a kinase-dead mutant. e, Quantitation of pMEK and pERK1/2 signals in 293T cells transected with the indicated MAP3K2 and SMYD3 constructs and treated as in (d). Data was generated from three independent biological replica. f, SILAC-based quantitative proteomic analysis of proteins that bind to MAP3K2-K260me0 and MAP3K2-K260me3 peptides. Data represent two independent experiments (forward and reverse direction). Proteins are plotted by their SILAC ratios in the forward (x axis) and reverse (y axis) SILAC experiments. Specific interactors of K260me0 reside in the lower left quadrant. The three PP2A complex components are highlighted in red. g, PP2R2A directly binds to MAP3K2 peptides encompassing amino acids 249-273 and this interaction is inhibited by K260 methylation. Immunoblots of peptide pulldowns as indicated with either recombinant proteins (top panel) or HeLa cytoplasmic extracts (middle panel) (based on two replica). The dot blot in the bottom panel shows equivalent amounts of peptides used for the experiments. h, Quantification of MUC5 positive lesions in caerulein-treated pancreata from Kras (n=5, each treatment) and Kras;Smyd3 (n=5, each treatment) mutant mice treated with the PP2A inhibitor cantharidin (iPP2A) (0.15mg/kg BID, IP) or vehicle control (see Extended Data Figure 10). *: p-value<0.05; **: p-value<0.01; ***: p-value<0.001; n.s.: not significant. (two-tailed unpaired Student's t-test). Data are represented as mean +/− SEM.
