Fig 3.
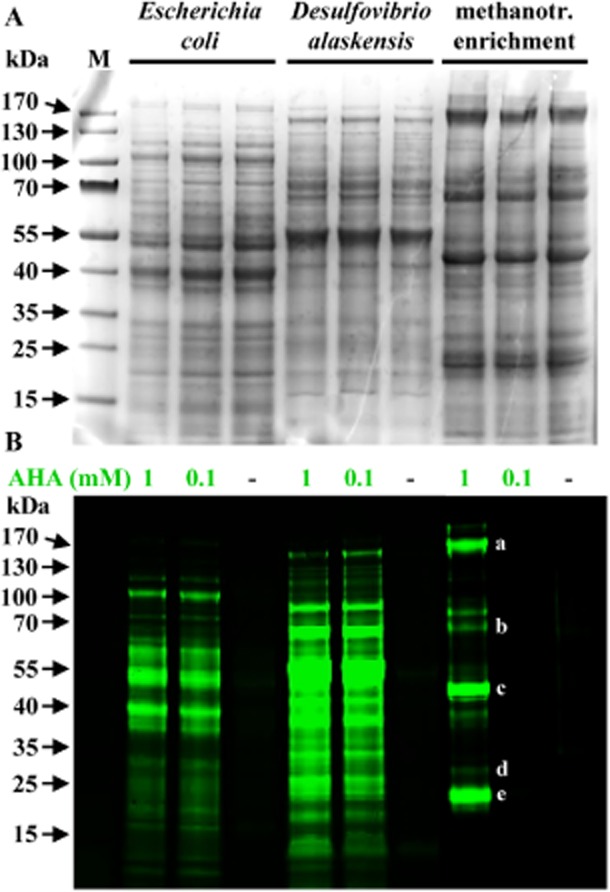
AHA does not interfere with the cellular machinery. Visualization of new proteins from cultures of E. coli, Desulfovibrio alaskensis and a methanotrophic enrichment. A. Coomassie-stained protein band patterns of cultures that had been incubated in the absence (−) or presence of AHA are indistinguishable from each other, demonstrating that AHA does not interfere with the translational machinery. B. Newly made proteins in the same gel are identified via BONCAT. Please note that the incubation time for the methanotrophic culture exposed to 100 μM AHA was too short to yield new proteins in amounts high enough to be detectable via in-gel fluorescence. At the individual cell level, AHA uptake can, however, be easily demonstrated (Fig. 2F). Some of the most intensely labelled bands were cut from the gel and analyzed via mass spectrometry. The 20 most abundant proteins from the excised bands included: (a) the two large subunits of RNA-polymerase (150.4 and 155.4 kDa); (b) a hypothetical protein (67.8 kDa) as well as two homologs of the large subunits of methanol dehydrogenase (66.6 and 68.3 kDa); (c) PmoB, 45.6 kDa; (d) PmoA (28.4 kDa) and PmoC (29.1 kDa); (e) a formaldehyde activating enzyme (17.8 kDa), superoxide dismutase (21.1 kDa), and D-arabino-3-hexulose 6-phosphate formaldehyde lyase (21.8 kDa). Letters a–e denote the bands consistent with the molecular weights of these proteins. kDa, kiloDalton; M, marker.
