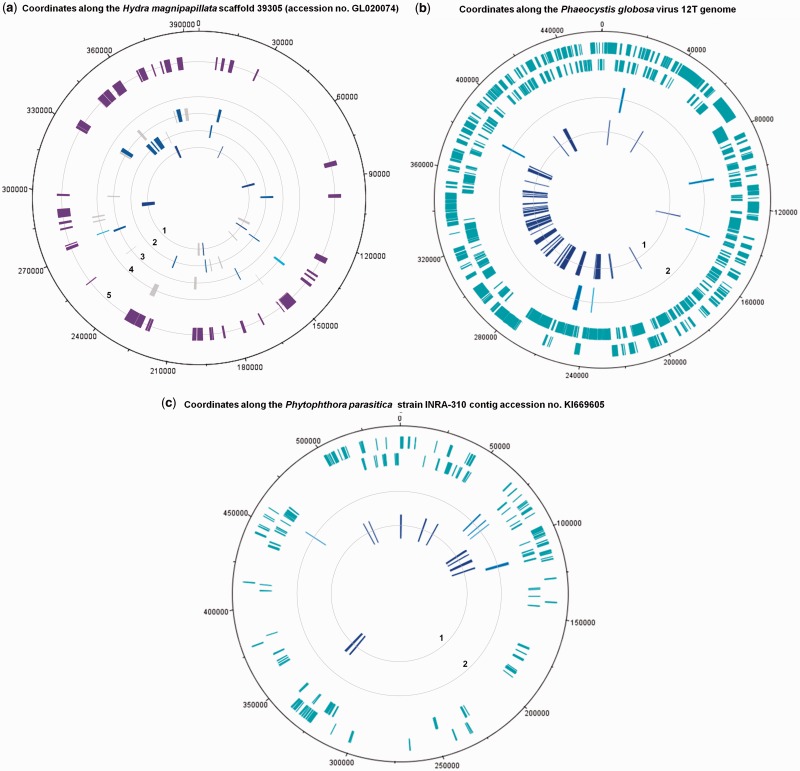
Fig. 2.—
Circular representations showing best and significant BLASTp hits for protein-coding predicted genes detected using GeneMarkS (Besemer and Borodovsky 2005) from a scaffold obtained from Hydra magnipapillata (GenBank accession number GL020074.1) (a) and for protein sequences downloaded from GenBank for the Marine Group II euryarchaeote SCGC AB-629-J06 (taxonomy ID: 1131268) draft genome (AQVM00000000) (b) and the contig accession no. KI669605 of Phytophthora parasitica strain INRA-310 (c). (a) Best hits corresponding to mimiviruses of lineages A, B, and C and distant mimiviruses (rings 1, 2, 3, and 4, respectively; blue) (Colson et al. 2012), and significant hits against a mimivirus at any rank among the 20 best hits (ring 5; purple) are mapped on the Hydra magnipapillata scaffold GL020074. Best hits corresponding to mimiviruses with a lower sequence coverage are colored in gray. (b) Best hits (ring 1) and other significant hits (ring 2) for predicted proteins from the Marine Group II euryarchaeote SCGC AB-629-J06 draft genome against the Phaeocystis globosa virus 12T gene repertoire (chosen because it provided the highest number of hits among the mimiviruses) are mapped on this viral genome. Outer rings indicate P. globosa virus 12T ORFs in sens (outer) and antisense (inner) orientations. (c) Best (ring 1) and other significant hits (ring 2) corresponding to a Megavirales member are mapped on the contig accession no. KI669605 of Phy. parasitica strain INRA-310. Outer rings indicate Phy. parasitica ORFs in sens (outer) and antisense (inner) orientations.