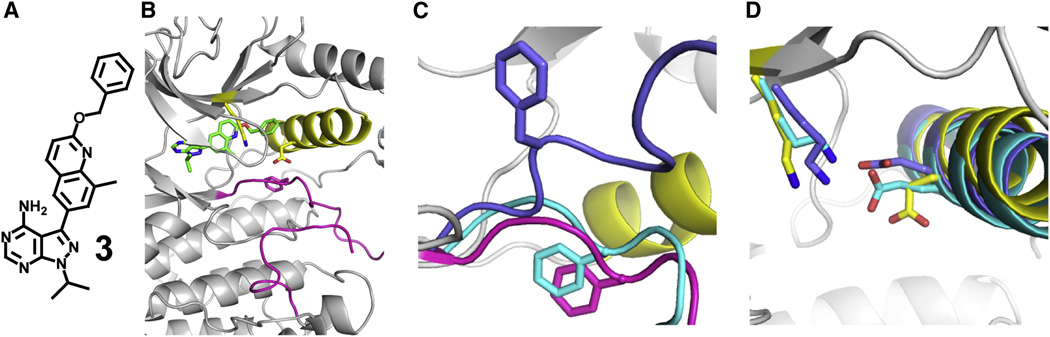
Figure 5. Erk2DS2 stabilized in a modified helix αC-out conformation.
A. Structure of ligand 3. B. Overall topology of 3-bound Erk2DS2. Helix αC is shown in yellow, the activation loop in magenta, and 3 in green. C. Superimposed structures of 3-bound Erk2DS2 (gray and yellow with magenta activation loop), SB203580-bound pentamutant Erk2 (cyan) (PDB ID: 1PME), and 2-bound Erk2DS2 (blue) (PDB ID: 4I5H). The DFG motif phenylalanine residues for all three structures are represented in stick form. The distances from the α-carbon of the phenylalanine residue in 2-bound Erk2DS2 from those of 3- and SB203580-bound Erk2 are 11.1 and 9.6 Å, respectively. D. Superimposed structures of 3-bound Erk2DS2 (gray and yellow), SB203580-bound pentamutant Erk2 (cyan) (PDB ID: 1PME), and 2-bound Erk2DS2 (blue) (PDB ID: 4I5H). Glu-69 and Lys-52 for all three structures are represented in stick form. The distances between these residues for 3-, 2-, and SB203580-bound Erk2 are 7.6, 2.7, and 2.7 Å, respectively. See also Figure S2 and Table S2.