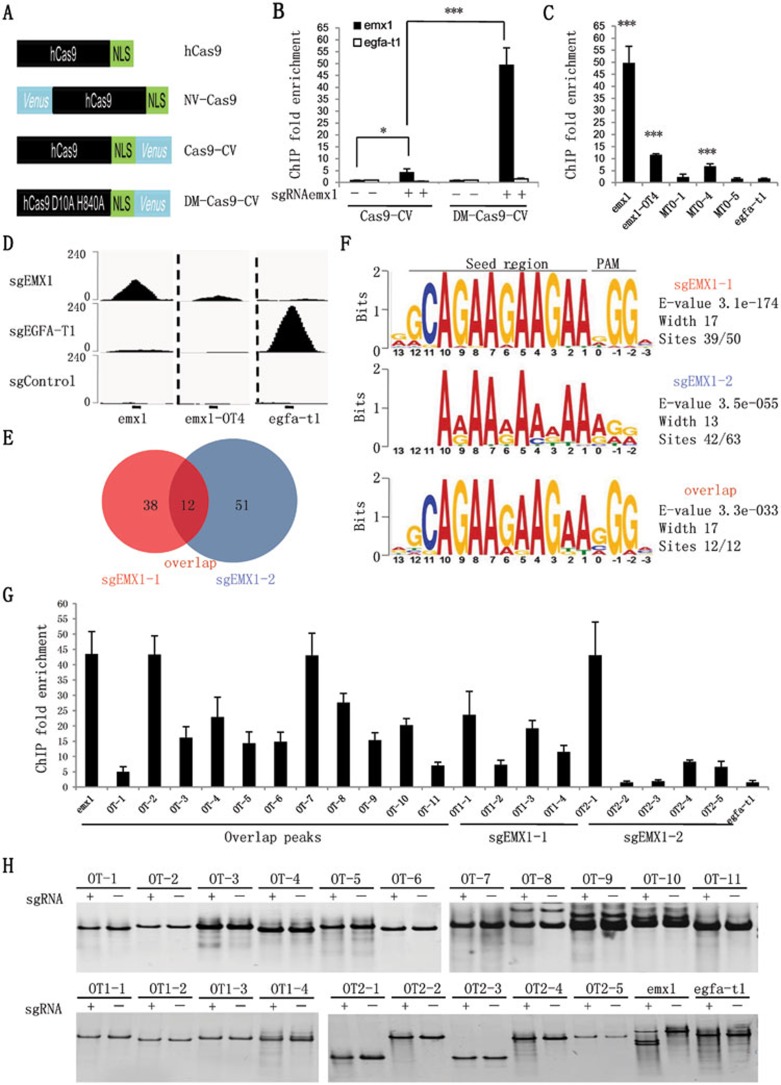
Figure 1.
GFP nanobody-based ChIP-seq analysis to unbiasedly identify genome-wide off-targets of CRISPR/Cas9 in human genome. (A) Schematic view of Cas9 constructs. Venus was fused to wild type hCas9 or hCas9 double mutant (D10A and H840A). (B) The sgRNA-specific in vivo binding of Cas9-CV and DM-Cas9-CV to endogenous emx1 and egfa-t1 loci. 293T cells were transfected with sgRNA for the emx1 locus or without sgRNA as control. The ChIP signals are divided by the no-sgRNA signals. (C) ChIP enrichment on emx1, previously identified emx1 off-target (emx1-OT4), control efga-t1, as well as other genomic loci containing sequences with 2-3 nucleotide mismatches in comparison to the original emx1 target sequence, was examined in 293T cells transfected with sgRNA for emx1. (D) ChIP signals (normalized read counts) around target and off-target sites in ChIP-seq libraries. (E) ChIP-seq peaks identified in sgEMX1 libraries. (F) Consensus binding motif analysis of peaks in sgEMX1 libraries. Sequence motifs were identified within 101 base pairs around peak summits using MEME. (G) Confirmation of ChIP-seq-identified peaks in sgEMX1 libraries by quantitative PCR. All 12 overlapped peaks as well as randomly selected peaks from the rest of individual sgEMX1 libraries were checked. (H) In vitro cleavage assay of identified binding off-target sites. Error bars indicate the standard deviation (STDEV). Student's t-test was performed (*P< 0.05, ***P< 0.0005). All experiments were replicated at least twice. See also Supplementary information, Figure S1 and Table S1.