Fig. 2.
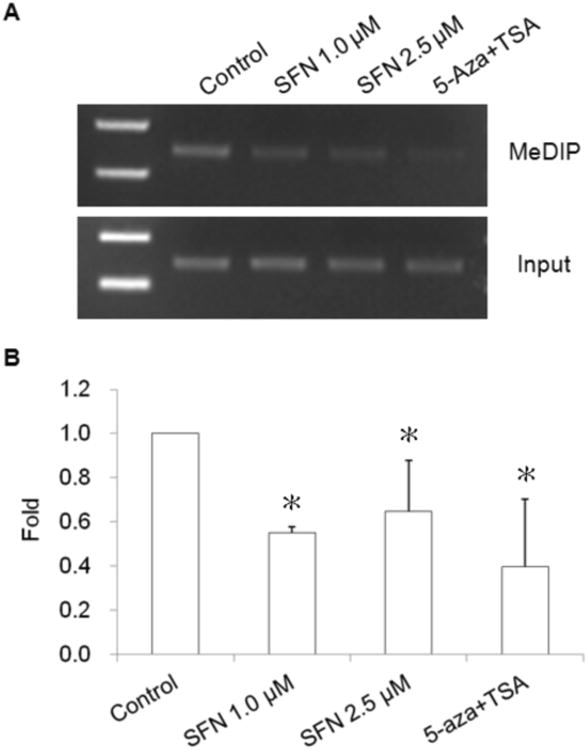
Effect of SFN on methylation of Nrf2 promoter region in TRAMP C1 cells using methylation DNA immunopredipitation (MeDIP) assay. Genomic DNA (10 μg) extracted from SFN or 5-aza/TSA treated TRAMP C1 cells were used for MeDIP analysis. Genomic DNA were sonicated, denatured and subjected to DNA immuniprecipitation (IP) with anti-methyl cytosine antibody. Then regular PCR (A) and qPCR (B) were performed to analyze the enrichment of methylated fragments using the primers covering the DNA sequence containing the first five CpGs in promoter region of the Nrf2 gene. (A) Regular PCR was performed to compare the immunoprecipitated DNA with their inputs, a representative result is shown from three independent experiments. (B) The enrichment of the MeDIP DNA was determined by qPCR on the basis of the standard curve from a serial dilution of the inputs. Relative methylated ratio was calculated by comparing with control group as 100% methylated DNA. Data are expressed as mean ± SD from four independent experiments. * Different from control, P < 0.05.
