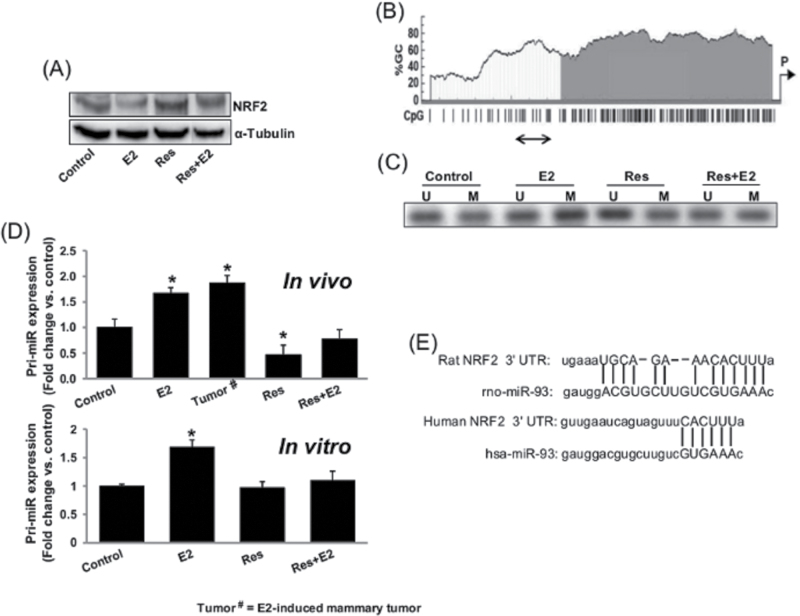
Fig. 4.
Resveratrol epigenetically regulates NRF2 expression. (A) Representative western blot showing NRF2 expression in MCF-10A cells treated with Res (50 µM) in the presence or absence of E2 (10nM) for 24h; (B) CpG-rich islands in NRF2 promoter as determined by MethPrimer software are shown. The promoter region amplified by methylation-specific PCR after bisulphite treatment is shown by a double-headed arrow; (C) Representative picture of agarose gel electrophoresis from methylation-specific PCRs with unmodified and bisulphite-modified genomic DNA from MCF-10A cells treated with Res (50 µM) in the presence or absence of E2 (10nM) for 24h is shown. U = PCR product with unmethylated DNA-specific primer pair; M = PCR product with methylated DNA-specific primer pair; (D) Real-time PCR analyses of pri-miR-93 expression in mammary tumors and in mammary tissues of rats treated with E2, Res and Res + E2 for 240 days and in MCF-10A cells after E2 (10nM), Res (50 µM) and Res + E2 treatment for 24h are carried out. The bar graphs represent a fold change in pri-miR-93 expression (mean ± SEM) in mammary tumors and in mammary tissues from at least four different animals compared with age-matched control mammary tissues (in vivo) and from at least five MCF-10A samples treated with E2 in the presence or absence of Res compared with vehicle-treated control samples (in vitro). * indicates P value of <0.05 compared with respective controls; and (E) Schematic of rat and human NRF2 mRNA 3′-UTR containing potential miR-93 binding site.