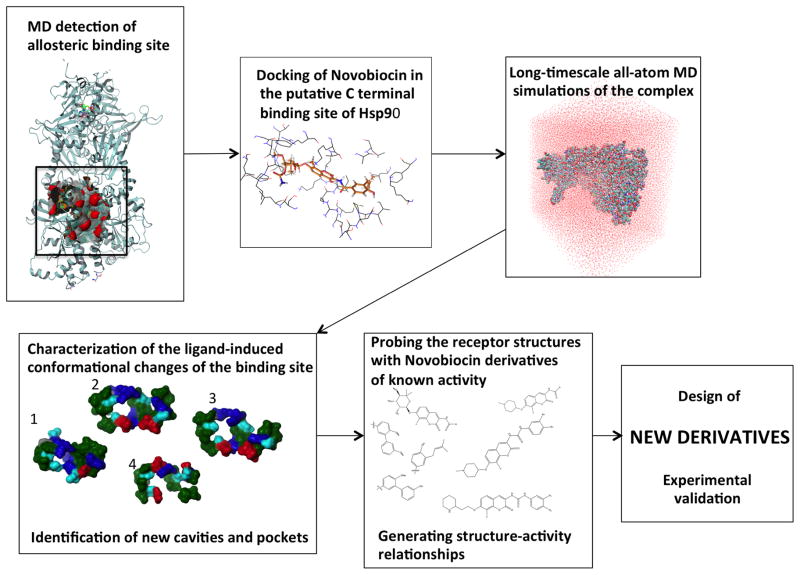
Figure 1. Strategy for the identification of new allosteric inhibitors using protein dynamics.
Identification of the putative C-terminal allosteric binding site from the analysis of the dynamics of the activated state of Hsp90 bound to ATP. Docking of Novobiocin, the first known lead to bind Hsp90-CTD, in this binding site and long-timescale all-atom MD simulations of the complex. The ensemble-based approach ensures the incorporation of receptor flexibility in the characterization of the ligand-induced conformational changes of the binding site. The new structures of the binding site have been probed with a series of analogues of known activity; this analysis allowed the identification of structure-activity relationships. This information has been exploited to design new derivatives with chemical modifications and functional groups that can reach into the cavities. Finally, selected hits have been tested experimentally