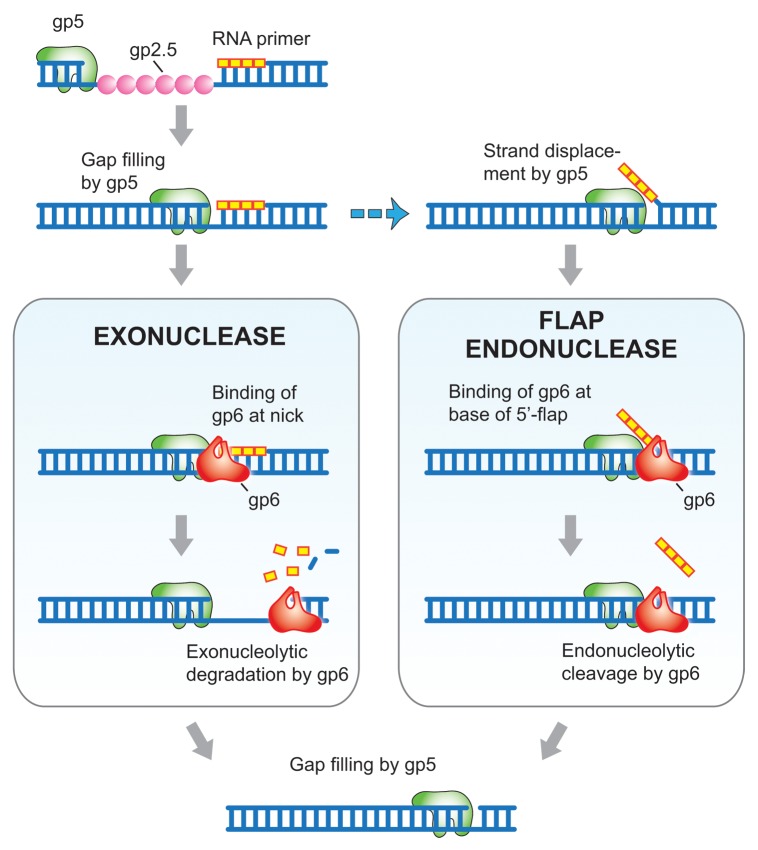
Figure 2. Model for the role of gp6 in RNA primer removal. T7 DNA polymerase (gp5 in green) fills the gap between Okazaki fragments, in the process displacing T7 single-stranded DNA binding protein (gp2.5 in pink) that is coating the single-stranded DNA template. When gp5 encounters the preceding Okazaki fragment two distinct pathways can occur: (i) The exonuclease activity of gp6 (red) degrades the RNA primer and continued hydrolysis results in the removal of some deoxyribonucleotides (left panel). (ii) Strand displacement synthesis by gp5, an event that can occur with exonuclease deficient gp5 or with exonuclease proficient gp5 in the presence of gp2.5, creates a flap containing the RNA primer (right panel). The flap endonuclease activity of gp6 cleaves the flap one nucleotide into the duplex, releasing the RNA primer. gp5 fills the gap resulting from removal of the flap and the resulting nick is sealed by T7 DNA ligase. DNA is indicated in blue and ribonucleotides are represented in yellow.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.