Figure 3.
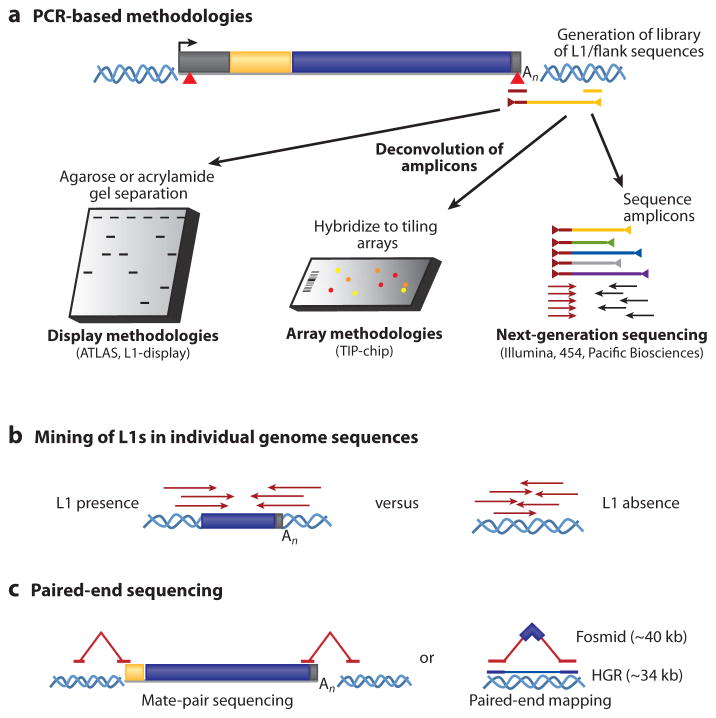
Methods to detect LINE-1-mediated polymorphic human retrotransposition events in individual genomes. Modifications of these assays can also be used to identify other polymorphic retrotransposons in human DNA. (a) Polymerase chain reaction (PCR)–based methodologies. PCR using primers specific to diagnostic sequence variants in the L1 (red triangles and the corresponding maroon primer) and arbitrary oligonucleotides or primers complementary to ligated linkers ( yellow line) can be used to amplify human-specific L1s and their associated flanking sequences (maroon/yellow line flanked by triangles). The amplicon libraries are then resolved using electrophoresis, and individual products are cloned and sequenced (left). Alternatively, the amplicons can be hybridized to genome tiling microarrays (center), or directly characterized using high-throughput sequencing methodologies (right). Abbreviations: ATLAS, amplification typing of L1 active subfamilies; TIP-chip, transposon insertion profiling by microarray. (b) Mining of L1s in individual genome sequences. Whole-genome sequences, comparative genomics, or mining trace sequence databases can discover dimorphic L1s in individual genomes that are absent from reference genome assemblies. (c) Paired-end sequencing. Mate-pair reads containing one sequence from a uniquely mapping portion of genomic DNA and one sequence from an L1 can be used to identify novel retrotransposons (left) in individual genomes. Paired-end sequencing of fosmid inserts with restricted size distributions (~40 kb) allows the discovery of novel ~6-kb insertions (right) as well as deletions and inversions relative to a reference sequence. Fosmids containing insertions can then be screened for the presence of human-specific L1s. Abbreviation: HGR, human genome reference sequence. These methods are also described in another recent review (182).