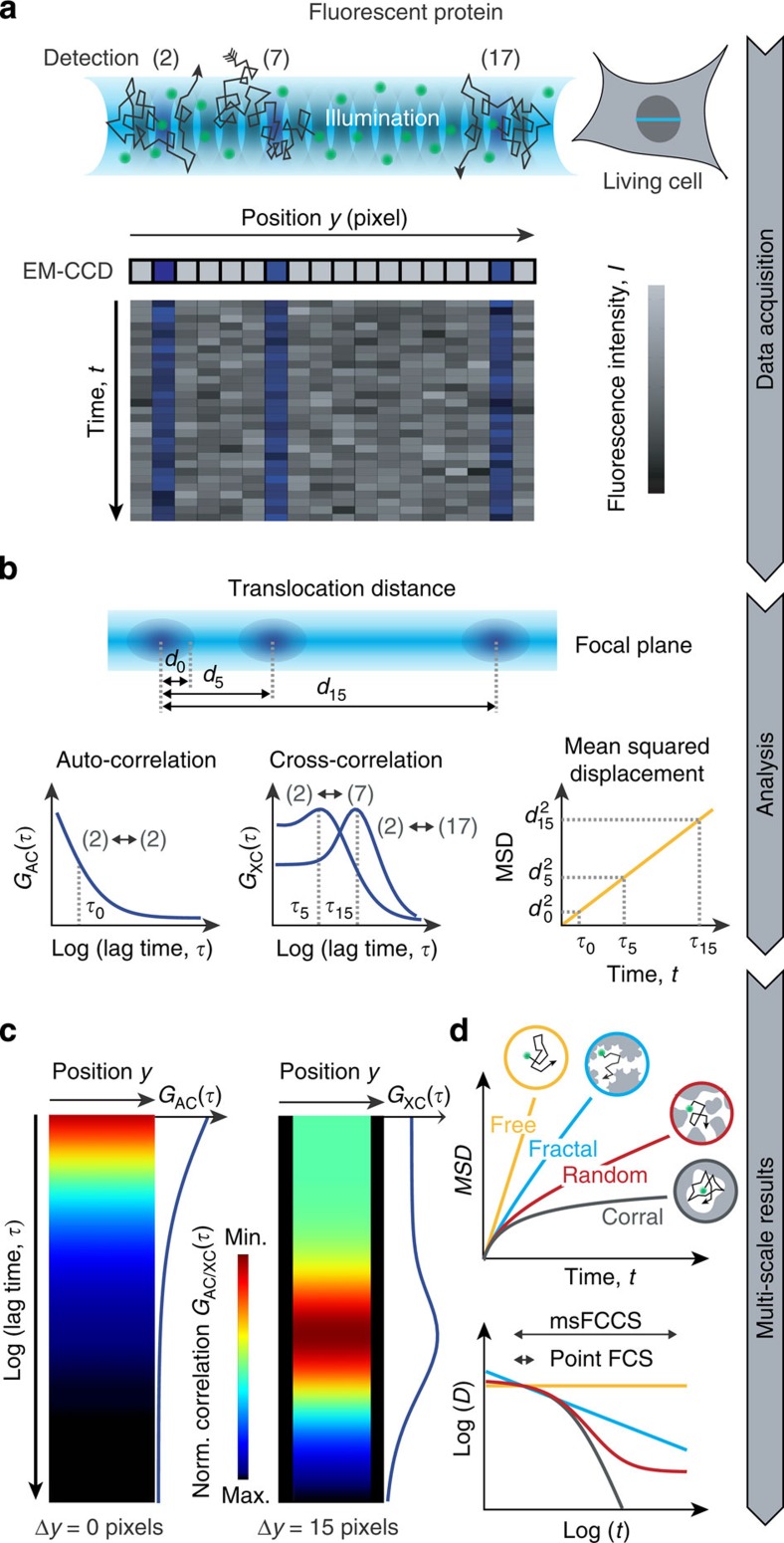
Figure 1. Parallelized acquisition of fluorescence signals for msFCCS analysis.
(a) Line illumination and parallelized multifocal fluorescence signal detection. Fluorescent proteins enter and leave the detection volumes by diffusion. The resulting local fluctuations in the fluorescence signal are detected on corresponding pixels of an EM-CCD camera array. (b) AC and XC analysis. By applying correlation analysis to the fluorescence signal recorded at a given detector pixel, AC curves can be calculated at every pixel position. Correlation of signals from spatially separated detection volumes are evaluated by computing XC curves (for example, signals of detection volumes 2 and 7 or detection volumes 2 and 17). These yield the MSD, the diffusion coefficient D and the concentration or molecule transmission rate as a function of the translocation distance dn or translocation time τn. (c) AC curves or XC curves for a constant distance between detection elements are acquired along the illumination line and are visualized in so-called correlation carpets, from which diffusion barriers can be identified. (d) The diffusion coefficients determined for different distances can be used to reconstruct the molecules’ MSD as a function of the diffusion time t. The time dependence of the diffusion coefficient reflects the nanostructure ‘seen’ by the diffusing protein. Norm, normalized.