Figure 3.
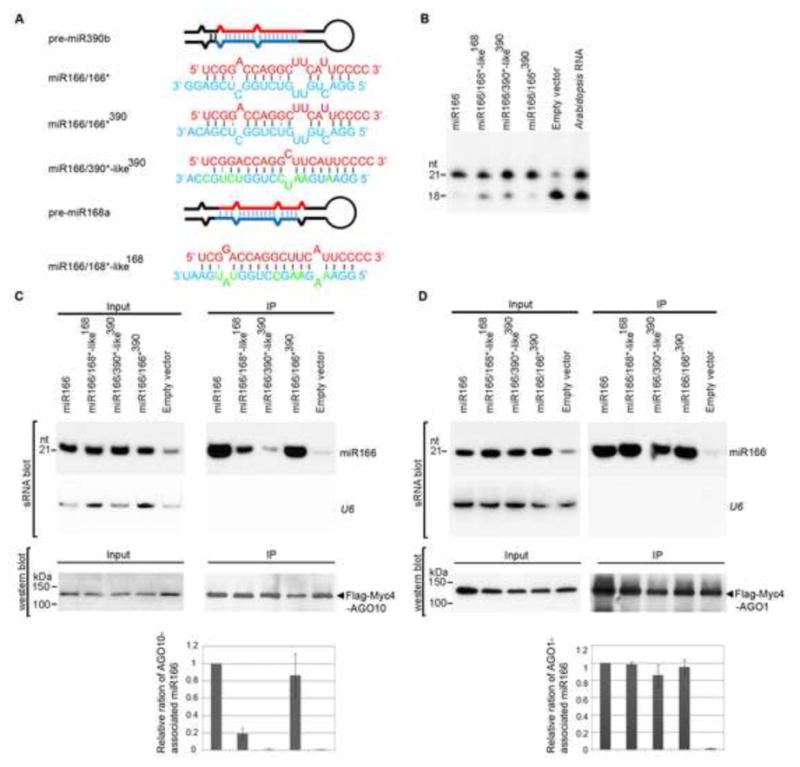
The internal structure of miR166/166* determines the specific AGO10-miR166 association. (A) Predicted foldbacks of pre-miR390b and -miR168a and chimeric precursors expressing miR166. miR166 (red) and its * strand (blue) are shown. The mutated nucleotides in the miR168*- and miR390*-like strands are shown in green. (B) Primer extension experiments were conducted with total RNAs prepared from N. bentha transfected with the indicated constructs. (C and D) Change of the miR166/166* structure dramatically decreased the loading of miR166 to AGO10 (C), but not to AGO1(D). Analyses of sRNA and western blots were conducted as in Figure 1E. The relative mean ratio of miR166/AGO10 (or AGO1) was normalized to that obtained with pre-miR166a with ±SD from five repeats (bottom panels). See also Fig. S3.
