Figure 5.
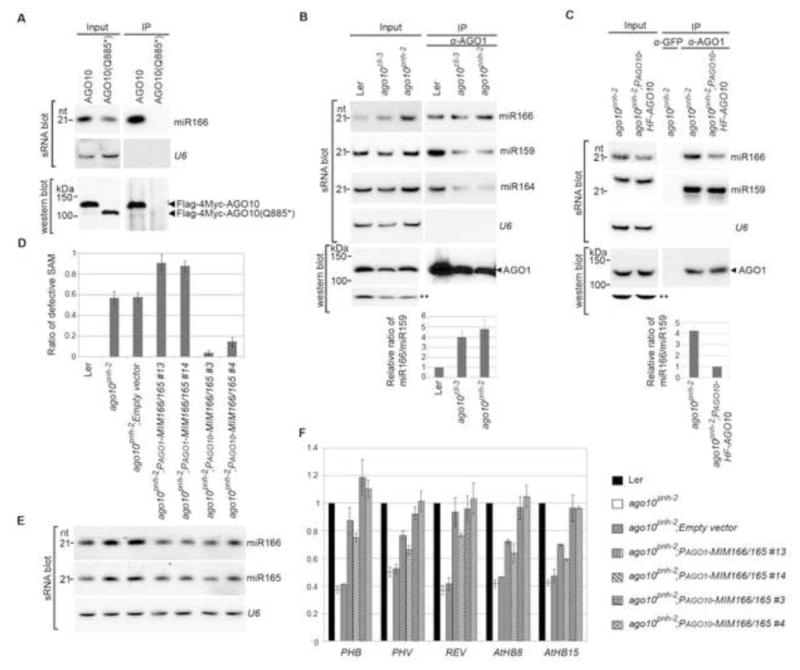
Sequestration of miR166/165 from expression domains of AGO10, but not AGO1, by target mimicry rescues the ago10pnh-2 phenotype. (A) AGO10 Q885* encoded by ago10pnh-2 did not bind to miR166 in N. bentha due to improper protein folding. Analyses of sRNA and western blots were conducted as in Figure 1E. (B and C) ago10 mutation resulted in a significant increase in miR166 binding by AGO1 in Arabidopsis. sRNA blot analyses were conducted with total RNA (input) and sRNA recovered from the AGO1 complexes (IP). Western blot assays were performed using an anti-AGO1 antibody. A cross-reacting band (**) served as a loading control. The relative signal ratio of miR166 to miR159 in AGO1 complexes was normalized to that obtained from the wild type Ler or the PAGO10-HF-AGO10 complemented lines with ±SD from three experiments (bottom panels). (D) Defective SAM was rescued in ago10pnh-2 plants by miR166/165 target mimicry expressed from the promoters of AGO10, but not AGO1. The ratios of defective SAM are shown as mean ±SD from three replicates (n > 200/each replicate). (E and F) Levels of miR166/165 and their target transcripts were measured by RNA blot assays (E) and real-time RT-PCR (F). The relative level of HD-ZIP transcripts was normalized to that in Ler plants with ±SD from four experiments. See also Fig. S5.
