Figure 6.
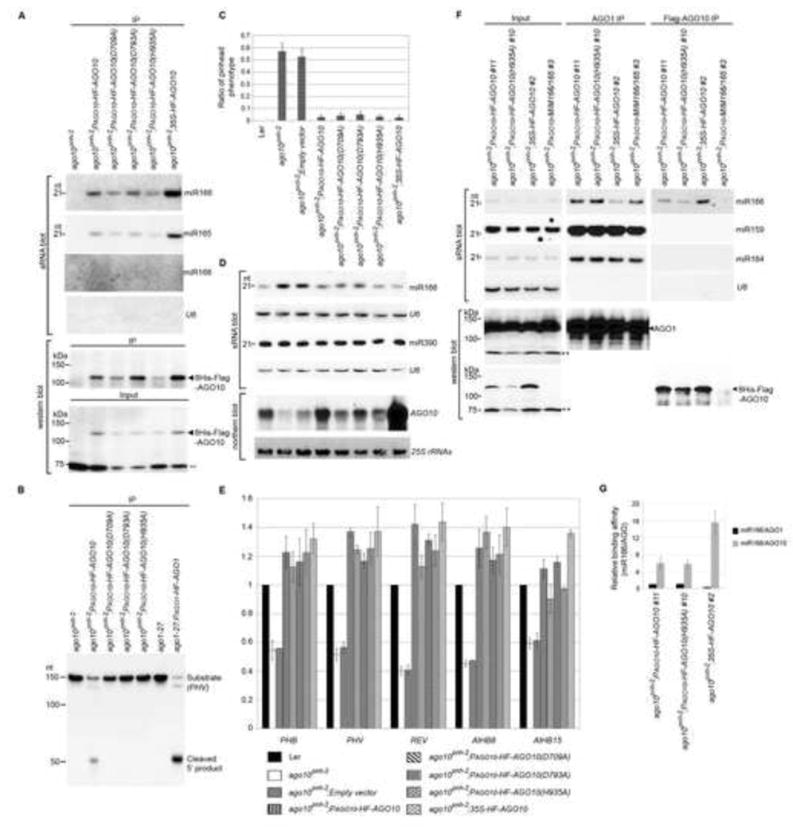
AGO10 rescues the ago10pnh-2 mutant by sequestering miR166/165 from AGO1. (A) AGO10 DDH mutants maintained miR166/165-binding capacity. Assays of sRNA blot and western blot (using anti-Flag antibody) were conducted as in Figure 1D. (B) RISC reconstitution assays of AGO10 and AGO10 DDH mutants. AGO1 was included as a positive control. (C) Non-catalytic AGO10 rescued the ago10pnh-2 mutant as efficiently as catalytic AGO10. The pinhead ratios are shown as mean ±SD from 16 lines (n > 200/line). (D and E) Levels of miR166/165, AGO10 and HD-ZIP III transcripts were measured by analyses of sRNA and northern blots (D) and real-time RT-PCR (E). The relative level of HD-ZIP transcripts was normalized as in Figure 5F.(F) AGO10 sequestered miR166 from AGO1. Analyses of sRNA blot and western blot (using the anti-AGO1 or anti-Flag antibody) were conducted as in Figure 1D. (G) The relative binding of miR166 by dual-tagged AGO10 was normalized to that of miR166/AGO1 isolated from ago10 pnh-2; PAGO10-HF-AGO10 plants with ±SD from three experiments. See also Figs. S5 and 6.
