Extended Data Figure 1. Maternal and embryonic gene expression patterns.
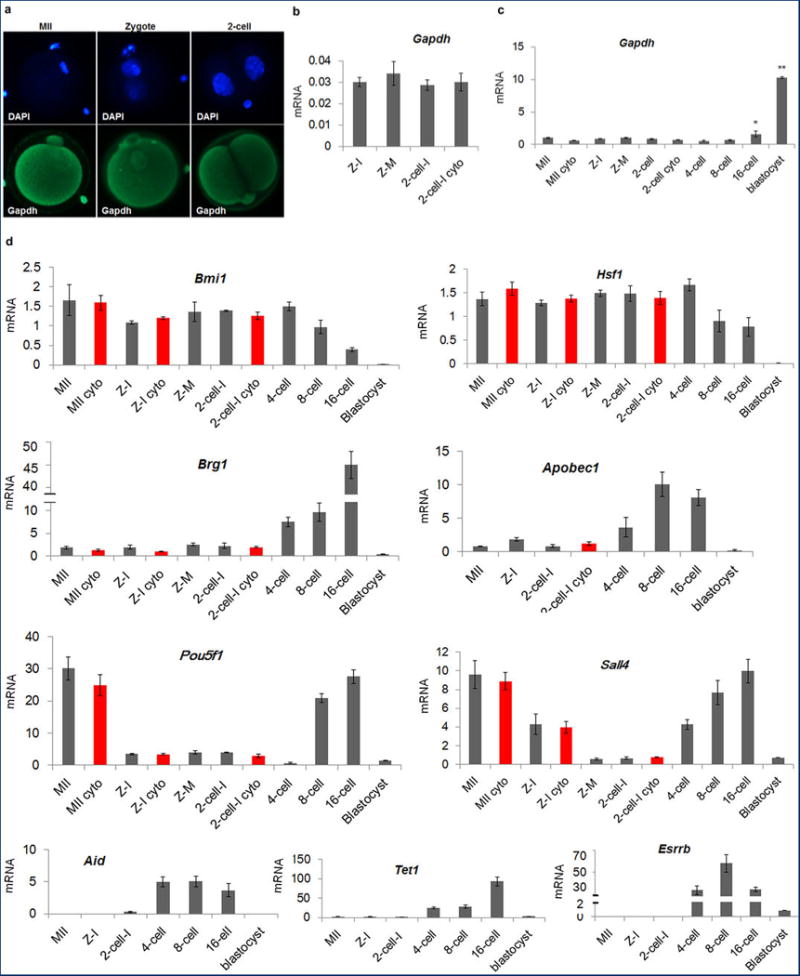
a, Immunocytochemical detection of Gapdh signal demonstrated even distribution in nuclei and cytoplasm in MII oocytes and interphase zygotes and 2-cell embryos. b, Expression of Gapdh normalized to β-actin. No significant differences were seen between intact and enucleated 2-cell embryos (4 replicates each containing pooled RNA from 5 embryos, p > 0.05). c, Gapdh expression was relatively low and constant with no significant differences seen until the 8-cell stage. Expression was increased in 16-cell embryos and blastocysts (3 replicates each containing pooled RNA from 10 embryos, *p < 0.05). d, Gene expression of the transcriptional regulators Bmi1, Hsf1, Brg1, Sall4 and Esrrb, transcription factor Pou5f1, and epigenetic factors Apobec1, Aid and Tet1 during mouse preimplantation embryo development. The level of expression for Brg1, Apobec1, Pou5f1, Sall4, Aid, Tet1 and Esrrb underwent dramatic increases at or after the 4-cell stage. The red bars indicate enucleated oocytes, zygotes or 2-cell embryos. No significant differences were observed between intact and enucleated counterparts (n=3 biological replicates, p < 0.05). Gene expression was normalized to Gapdh. Error bars indicate average ± s.d., MII = Metaphase II arrested oocyte, Z =Zygote, I = Interphase, M = mitotic, Cyto = Cytoplast.
