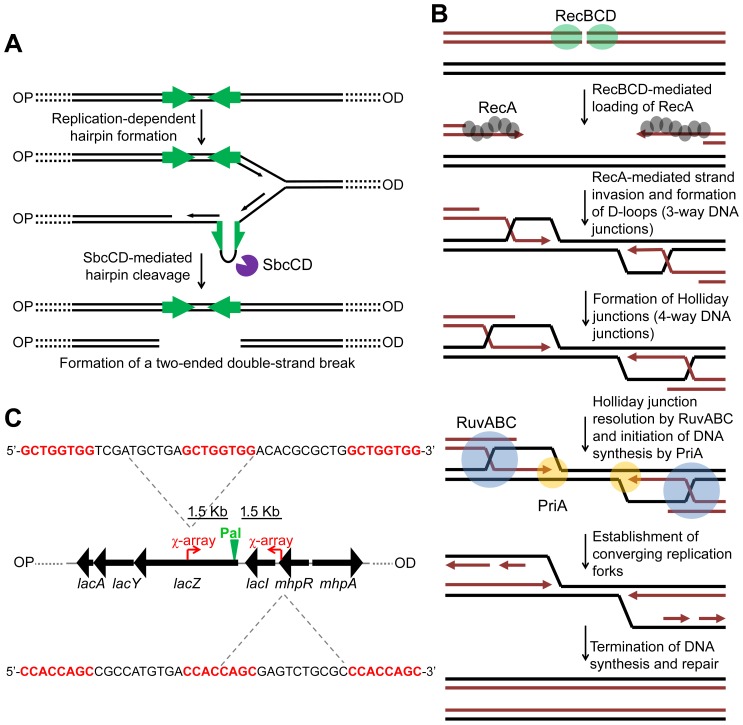
Figure 1. Making and repairing a site-specific DNA double-strand break in the E. coli chromosome.
(A) SbcCD-mediated cleavage of a 246 bp interrupted palindrome inserted into the chromosomal lacZ gene. During replication, the palindrome becomes transiently single-stranded on the lagging-strand template. This allows it to form a DNA hairpin that is cleaved by SbcCD, generating a two-ended DSB. OP and OD indicate origin-proximal and origin-distal sides of the break, respectively. The palindrome is highlighted by green arrows. (B) RecBCD-mediated HR. The ends of the break are processed by RecBCD to generate 3′ ssDNA overhangs coated in RecA. RecA searches the genome for a homologous DNA sequence and catalyses strand-invasion. This forms a D-loop and HJs. The D-loop is acted upon by the replisome assembly factor, PriA, which initiates DNA synthesis. The HJs can be acted upon by RuvABC, branch-migrated and resolved. This generates two converging replication forks, which, upon convergence, terminate the repair process. (C) Map of the lacZ region of the E. coli chromosome illustrating the position and sequence of two 3x χ arrays that have been inserted 1.5 Kb either side of the palindrome in order to stimulate recombination in close proximity of the DSB. The 8 bp χ recognition sequence, highlighted in red, is repeated three times. OP and OD indicate origin-proximal and origin-distal sides of the break, respectively. Pal represents the position of the palindrome.