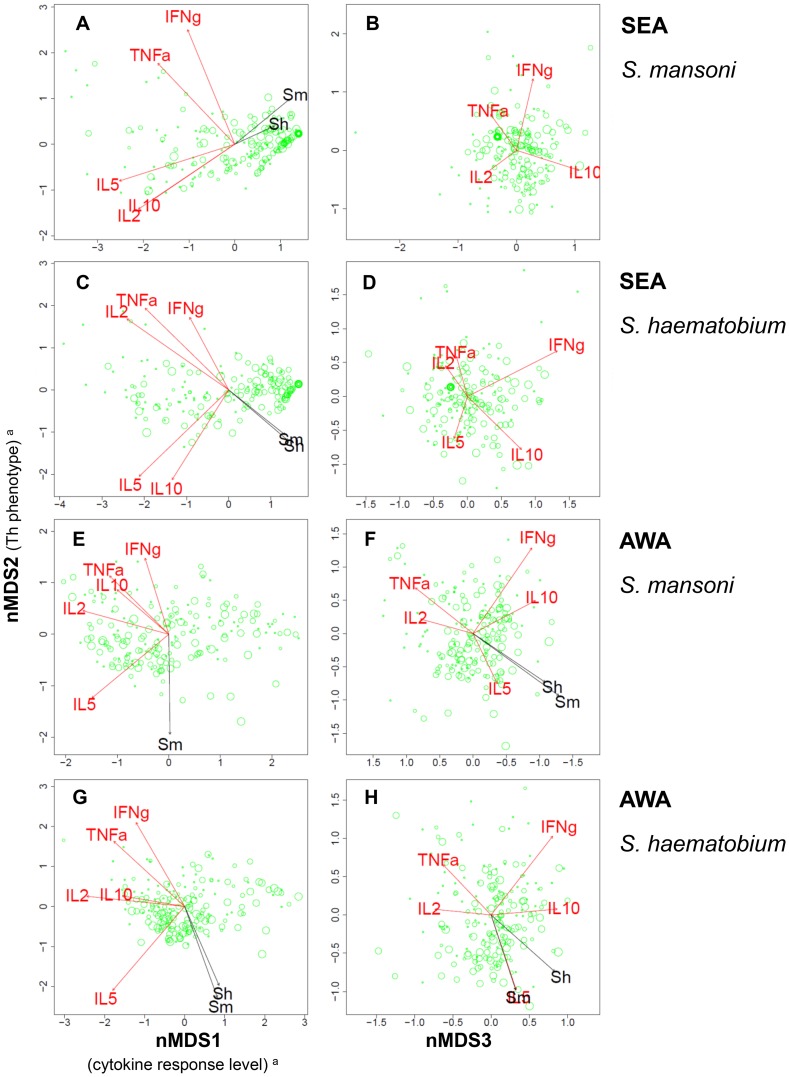
Figure 1. Variation in Schistosoma antigen-induced cytokine responses in relation to Schistosoma infection intensity.
Each three-dimensional (3D) nMDS ordination is represented in two 2D planes (Supporting Information S1). Left and right panels represent the 1st and 2nd, and 2nd and 3rd dimensions, respectively. Panels A and B show the S. mansoni egg antigen (SEAm)-induced cytokine profile, Panels C and D that of S. haematobium SEA(h), Panels E and F that of S. mansoni adult worm antigens (AWAm), and Panels G and H show S. haematobium AWA(h)-induced cytokine profiles. Green dots represent individuals. Distances between dots approximate the rank order of dissimilarities in cytokine profiles between the respective individuals with stress values (i.e. discrepancies) of 0.051 for SEAm, 0.041 for SEAh, 0.058 for AWAm, and 0.061 for AWAh. Red arrows indicate linear gradients of normalized net cytokine responses on which the nMDS is based. Green dot sizes are proportional to individual values of normalized infection intensity of S. mansoni (for simplicity dots were only labelled with S. mansoni (not S. haematobium) infection intensity). Black arrows indicate linear gradients of post hoc fitted normalized infection intensity of S. mansoni (‘Sm’) and S. haematobium (‘Sh’). The length of the arrows is proportional to the goodness of fit onto the cytokine profile within one 2D plane, but lengths cannot be compared between cytokine and infection intensity arrows. Arrows are only depicted if their fit was significant at the level of p = 0.05 in 3D ordinations (see Table 4), as well as in the respective 2D planes. In Panel H, the arrows of IL-5 response and S. mansoni infection intensity are overlapping and their labels are therefore illegible. aThe biological a posteriori interpretation of nMDS1 (left x-axis) and nMDS2 (y-axis) were added between brackets on the axis labels, but nMDS3 (right x-axis) could not be interpreted.