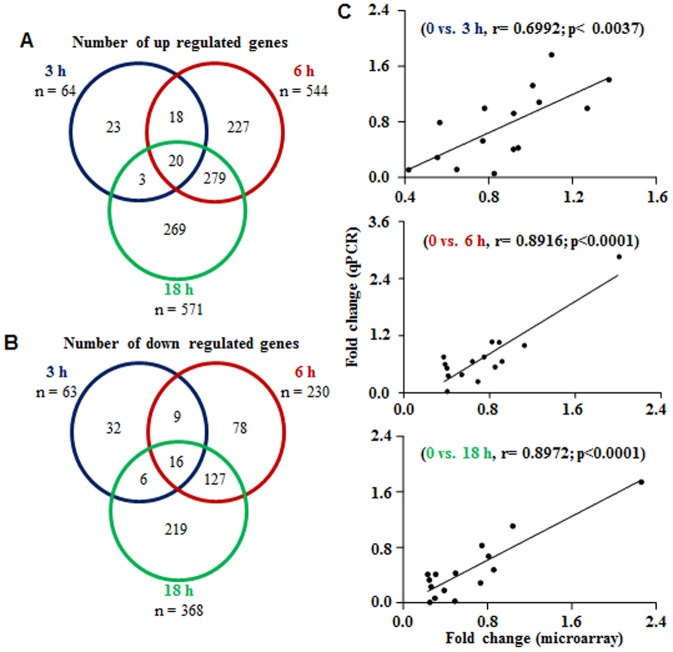
Figure 1. Schematic representation of differentially expressed genes in the CL post PGF2α treatment.
(A and B) Venn diagrams representing the number of differentially expressed genes identified after microarray analysis of CL tissues collected before (0 h) and at different time points post PGF2α treatment. Data analyzed by Bioconductor analysis tool employing ≥2 fold change cut-off and statistical filters with Benjamini and Hochberg correction factor for false discovery rate. The comparison of total number of differentially expressed up (A) and down (B) regulated genes found common between 0 vs. 3 h (blue circle) and 0 vs. 6 h (red circle), as well as between 0 vs.18 h (green circle) post PGF2α treatment are presented. (C) Correlation analysis between expression ratios obtained from microarray and qPCR analyses of few genes post PGF2α treatment (n = 15 genes/time point). Linear regression analysis was performed for selected differentially expressed genes using qPCR fold change in expression values (2−ΔΔCT; Y-axis) with fold change expression values obtained by microarray analysis (X-axis). The r value, correlation coefficient, generated for the theoretical line of best fit (represented as solid line in each panel) and the p value indicate the significance of the correlation as determined by ‘F’ test.