Abstract
Acetylation of histones is a key regulatory mechanism of gene expression in eukaryotes. GcnE is an acetyltransferase of Aspergillus nidulans involved in the acetylation of histone H3 at lysine 9 and lysine 14. Previous works have demonstrated that deletion of gcnE results in defects in primary and secondary metabolism. Here we unveil the role of GcnE in development and show that a ∆gcnE mutant strain has minor growth defects but is impaired in normal conidiophore development. No signs of conidiation were found after 3 days of incubation, and immature and aberrant conidiophores were found after 1 week of incubation. Centroid linkage clustering and principal component (PC) analysis of transcriptomic data suggest that GcnE occupies a central position in Aspergillus developmental regulation and that it is essential for inducing conidiation genes. GcnE function was found to be required for the acetylation of histone H3K9/K14 at the promoter of the master regulator of conidiation, brlA, as well as at the promoters of the upstream developmental regulators of conidiation flbA, flbB, flbC, and flbD (fluffy genes). However, analysis of the gene expression of brlA and the fluffy genes revealed that the lack of conidiation originated in a complete absence of brlA expression in the ∆gcnE strain. Ectopic induction of brlA from a heterologous alcA promoter did not remediate the conidiation defects in the ∆gcnE strain, suggesting that additional GcnE-mediated mechanisms must operate. Therefore, we conclude that GcnE is the only nonessential histone modifier with a strong role in fungal development found so far.
Keywords: Gcn5, GcnE, histone acetylation, SAGA, Aspergillus, conidiation, asexual development, brlA, fluffy genes
CHROMATIN rearrangements are associated with the transcriptional regulation of gene expression in eukaryotes. For example, facultative heterochromatin can be associated with the transcriptionally active or silent states of developmentally regulated loci (Grewal and Jia 2007). This is achieved in part through histone post translational modifications (PTM), which play a very important role in the control of these active or silent chromatin states. Histone modifications include acetylation, methylation, phosphorylation, and ubiquitination at different positions of the histone proteins. In particular, acetylation of lysine 9 or lysine 14 in histone H3 has been associated with activation of transcription. Acetylation of histones plays two roles in the regulation of transcription: it alters the physical properties of the histone–DNA interaction, and it also provides a frame for the binding of bromodomain proteins that remodel the chromatin and regulate gene expression (Spedale et al. 2012). These modifications regulate the nucleosome positioning at the gene promoters and the recruitment of the regulatory proteins. One of these modifiers, the SAGA complex, is responsible for the acetylation of several lysine residues in the N-terminal region of histones, particularly histone H3 lysine 9 (H3K9) and histone H3 lysine 14 (H3K14) (Kuo et al. 1996). The SAGA complex is a multimeric protein association with several subunits including Ada2p, Ada3p, Spt3p, and Tra1p (Grant et al. 1997; Spedale et al. 2012), where Gcn5p is the subunit with the histone acetyltransferase (HAT) catalytic activity (Grant et al. 1997). The SAGA complex is implicated in several functions related to transcription, such as transcription initiation and elongation, histone ubiquitination, and interactions of TATA-binding proteins. In addition, SAGA has also been implicated in messenger RNA (mRNA) export in yeasts and Drosophila (Rodriguez-Navarro et al. 2004; Kurshakova et al. 2007). In Saccharomyces cerevisiae, the SAGA complex is involved in the transcriptional regulation of 12% of the yeast genome. Approximately, a third of that 12% of the yeast genome is downregulated and two-thirds are upregulated in ΔGCN5 cells (Lee et al. 2000), implying a direct or indirect negative role of Gcn5p. Interestingly, a high proportion of genes regulated by SAGA are upregulated during the responses to environmental stresses (such as heat, oxidation, and starvation) (Huisinga and Pugh 2004). The SAGA complex is also present in metazoans, where it has diverged and evolved into four different complexes (two SAGA and two ATAC complexes), while lower eukaryotes, such as yeasts and other fungi, contain one single SAGA complex. It was hypothesized that this evolution into a diverse set of complexes is involved in cellular specialization during development and homeostasis in metazoans (Spedale et al. 2012). The SAGA and ATAC complexes participate in the regulation of genes in response to intracellular and extracellular signals: protein kinase C signaling, response to osmotic stress, UV-induced DNA damage, arsenite-induced signaling, endoplasmic reticulum stress, and nuclear receptor signaling (Spedale et al. 2012). Likewise, plants also have multiple HATs. In Arabidopsis, AtGCN5 is involved in many developmental processes (Servet et al. 2010).
Although elegant experimental approaches using Neurospora crassa as a model system have significantly contributed to general concepts of DNA methylation, genome defense, and heterochromatin formation (Tamaru and Selker 2001; Freitag et al. 2002, 2004; Honda et al. 2010; Rountree and Selker 2010), studies on transcriptionally related chromatin rearrangements and histone modifications are still scarce in filamentous fungi, a broad group of ecologically, industrially, and clinically important organisms. In N. crassa, the transcriptional activation of the light-inducible gene al-3 requires the acetylation of histone H3K14 by a homolog of Gcn5p, NGF-1 (Grimaldi et al. 2006), and in Aspergillus nidulans the SAGA/ADA complex is involved in the acetylation of H3K9/K14 at the prnD-prnB bidirectional promoter during inducing conditions, but increased levels of H3K9ac/K14ac are not required for transcription (Reyes-Dominguez et al. 2008). Georgakopoulos et al. (2013) reported the delineation of the A. nidulans SAGA complex with a combined proteomics and bioinformatics approach revealing a high conservation with the yeast SAGA complex except for the deubiquitination–H2B–Ub complex. Only recently, the relevance of chromatin-based silencing of secondary metabolite gene clusters was recognized in several Aspergillus and Fusarium species (Shwab et al. 2007; Bok et al. 2009; Lee et al. 2009; Reyes-Dominguez et al. 2010; Strauss and Reyes-Dominguez 2011). For example, it has been demonstrated that acetylation of histone H3 is required for the synthesis of secondary metabolites in A. nidulans (Reyes-Dominguez et al. 2010; Nützmann et al. 2011; Bok et al. 2013; Nützmann et al. 2013). Reduction of heterochromatin marks leads to higher secondary metabolite production in Aspergillus and Fusarium species (Reyes-Dominguez et al. 2010, 2012), and it has also been found that it de-represses silent clusters, leading to the production of novel metabolites (Bok et al. 2009). In addition, adverse metabolic and morphologic effects are also observed in histone modifier mutants, for example, deletion of the histone H3K9 methyltransferase clrD in Aspergillus fumigatus resulted in reduced radial growth and also delayed transcriptional activation of brlA and conidiation (Palmer et al. 2008).
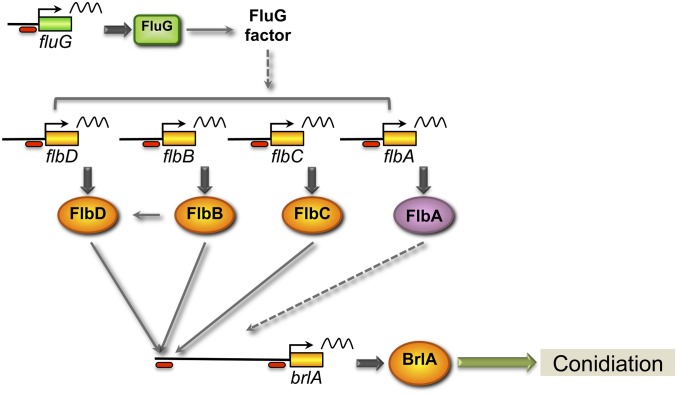
Asexual reproduction, also called conidiation, results in the formation of mitotic propagules (conidia), which are the infectious particles for pathogenic filamentous fungi. Conidiation is the most common and proliferative reproductive mode in filamentous fungi. For this reason, conidiation has been extensively studied in A. nidulans for several decades (for recent reviews see Etxebeste et al. 2010; Park and Yu 2012; Krijgsheld et al. 2013). Conidiation is controlled by a central regulatory pathway (Figure 1), encompassing three transcriptional activators: BrlA, AbaA, and WetA (see reviews by Adams et al. 1998; Yu et al. 2006). The first component in this regulatory cascade, BrlA, is essential to drive conidiation (Adams et al. 1988). brlA expression is silent during vegetative growth, and its expression during conidiation is controlled by a number of genes, including the fluffy genes. Deletion of any of the fluffy genes gives a typical fluffy phenotype with cotton-like colonies, lack of normal conidia, and reduced levels of brlA expression (Adams et al. 1998; Yu et al. 2006). There are six fluffy genes: fluG and flbA–E. fluG encodes a protein similar to bacterial glutamine synthetases (Lee and Adams 1994), and the FluG protein is responsible for the synthesis of the extracellular factor dehydroaustinol that, in conjunction with the orsellinic acid derivative diorcinol, induces conidiation (Rodriguez-Urra et al. 2012). FluG works upstream of the flbA-E genes (Yu et al. 2006). Flb genes operate in three parallel routes in A. nidulans to regulate the expression of brlA upon induction of conidiation. FlbA is a repressor of the G-protein signaling, which participates in a protein kinase A-dependent pathway to promote filamentous growth and to inhibit conidiation (Yu et al. 1996). FlbE interacts with FlbB at the fungal tip and is required for proper activation of FlbB (Garzia et al. 2009). FlbB is a bZip transcription factor that activates the transcription of flbD, a cMyb-type regulator. Then, both FlbB and FlbD jointly activate the transcription of brlA (Garzia et al. 2010). FlbC is a putative C2H2 Zn finger protein that constitutes a third route for the regulation of brlA expression (Kwon et al. 2010). These fluffy genes are expressed in vegetative mycelium and are able to respond to intracellular stimuli to induce a coordinated activation of the master regulator brlA (Etxebeste et al. 2010).
Figure 1.
Simplified model of the genetic regulation of conidiation. Only some of the regulators studied in this work are shown for clarity. FluG is responsible for the synthesis of an extracellular factor that induces the rest of the fluffy genes in the three parallel routes. FlbE (not shown) interacts and activates FlbB. FlbB and FlbD are transcription factors that jointly bind to the promoter of brlA, activating its transcription. FlbC is another transcription factor activating the expression of brlA. FlbA is a regulator of G-protein activity that positively regulates the transcription of brlA. Activation of brlA is necessary and sufficient to induce conidiation. Ovals indicate the promoter regions, and in front of brlA correspond to the two sites analyzed by ChIP.
Previous work noted that deletion of the SAGA subunits gcnE or adaB in A. nidulans resulted in strongly reduced conidiation, while not affecting the activation (but repressibility) of the proline utilization genes prnD-prnB, which are transcribed divergently from a bidirectional promoter (Reyes-Dominguez et al. 2008). Interestingly, Georgakopoulos et al. (2012) also found a lack of acetate repressibility in a SAGA-defective mutant. Here, we clarify the role of GcnE in the control of fungal development.
Materials and Methods
Strains, media, and culture conditions
The A. nidulans strains used in this study are listed in Supporting Information, File S1, and Table S1. Strains were grown in complete or minimal media containing the appropriate supplements (Cove 1966). Glucose was used as the carbon source and ammonium nitrate was used as the nitrogen source. In the brlA-overexpressing experiments, threonine and fructose were used as carbon sources to overexpress the brlA gene, and glucose was used to repress the brlA gene under the control of the promoter of the alcA gene. Ammonium tartrate was used as nitrogen source. Strains were obtained following standard procedures (Pontecorvo et al. 1953). Trichostatin A, butyric acid, and valproic acid were purchased from Sigma and used as histone deacetylase (HDAC) inhibitors at a concentration of 5 µM.
RNA isolation and real-time RT-PCR
Isolation of RNA and quantification of mRNA were performed as previously described (Ruger-Herreros et al. 2011). Briefly, mycelia (100–200 mg) were disrupted in 1 ml of TRI reagent (Sigma) with 1.5 g of zirconium beads by using a cell homogeneizer (FastPrep-24, MP Biomedicals). Cell debris was removed by centrifugation, and RNA samples were further purified using the NucleoSpin RNA II Kit (Macherey-Nagel).
The primers employed for real-time RT-PCR are detailed in Table S2. Real-time RT-PCR experiments were performed in triplicates (technical replicates) in a LightCycler 480 II (Roche) by using the One Step SYBR PrimeScript RT-PCR Kit (Takara Bio Inc.). The fluorescent signal obtained for each gene was normalized to the corresponding fluorescent signal obtained with the β-tubulin gene benA to correct for sampling errors. Expression data are the average of at least three independent biological replicates.
Microarray experiment
Strains were grown in complete liquid medium for 18 hr at 37°, and then conidiation was induced by transferring the vegetative cultures to complete solid media. Strains were further grown for 10 hr at 37°. Samples were immediately frozen in liquid nitrogen upon harvesting and stored at –80° until processing. RNA was isolated from strains grown in liquid or solid media as previously reported (Schinko et al. 2010). RNA samples were quality controlled with the Agilent 2100 Bioanalyzer using the RNA 6000 Nano Kit. For each array, 1 µg of total RNA was labeled with MessageAmpTMII-Biotin Enhanced RNA Kit (Ambion) according to the manufacturer’s instructions. Hybridizations were done automatically for 16 hr at 45° using the GeniomRT Analyzer. The array underwent a stringent wash. Following the labeling procedure, a microfluidic-based primer extension assay was performed. This assay utilized the bound mRNAs as a primer for an enzymatic elongation with labeled nucleotides. The elongation was done with Klenow Fragment and biotinylated nucleotides at 37° for 15 min. Finally, the array was washed automatically and detection was achieved with streptavidin–phycoerythrin using a Cy3 filter set in a GeniomRT Analyzer. Three independent biological replicates were obtained for each sample.
Analysis of microarray data
The experimental dataset is deposited in the Gene Expression Omnibus database (accession no. GSE48426). For the four conditions, further data analysis was performed in the Bioconductor R (http://www.bioconductor.org/). Raw intensity values were imported into R for statistical analysis using the Limma package (Smyth 2005). First, we carried out a global background subtraction; i.e., for each array, the global background was computed and subtracted from the measured intensity. To account for variations between the hybridized arrays, variance stabilizing normalization (VSN) was used. The normalized data were thereby transformed to a so-called generalized log scale. Thus, the fold quotients were also calculated on a log scale (qmedian). To provide estimates of the fold quotients, we utilized the exponential function. This was roughly equivalent to using the natural logarithm instead of log2 (log-qmedian).
For the detection of differentially regulated genes between vegetative growth and conidiation in the wild-type or the mutant strain, the Empirical Bayes test statistics (Smyth 2005) was used. The raw P-values were adjusted for multiple testing to control the false discovery rate by using the Benjamini–Hochberg (BH) method (Benjamini and Hochberg 1995) with a cutoff of adjusted P-value of <0.05. Under this criterion, all selected genes showed a minimum log-qmedian < −0.7 or >0.7 (where a log-qmedian of 0.7 was roughly equivalent to a twofold change).
To study the effect of the different factors (mutant, wild type, vegetative growth, and conidiation) on gene expression, we performed the ANOVA test of the normalized intensities using the Babelomics 4 suite (Medina et al. 2010). Differentially expressed genes were selected using a cutoff of adjusted P-values (BH method) of 0.05. The normalized intensities of the genes selected by the ANOVA test were used for PC analysis and clustering of the 12 samples (under four different conditions with 3 replicate samples each). The PC analysis was performed with PAST (Hammer et al. 2001). Cluster analysis was performed by centroid linkage clustering of the euclidean distances in Eisen’s modified Cluster 3.0 (de Hoon et al. 2004). Gene Ontology (GO) analysis was performed with the GO Term Finder tool at the Aspergillus Genome Database site (http://www.aspergillusgenome.org) (Arnaud et al. 2010).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were carried out as previously described (Reyes-Dominguez et al. 2008) with primers listed in Table S2. DNA was immunoprecipitated with antibodies recognizing acetylated K9 and K14 of histone H3 (Millipore ab 06-599) or the C terminus of histone H3 (Abcam ab1791). For each sample, the absolute amount of the specific DNA fragment in the immunoprecipitated sample was divided by the amount of this fragment in the sample before precipitation (normalizing to input DNA). The values shown are the averages of at least three biological repetitions. Standard errors are indicated.
Scanning electron microscopy
Strains were grown on complete solid medium for 7 days at 37°. Samples were prepared for electron microscopy as previously reported (Canovas et al. 2011) with some modifications. Briefly, excised cubes of agar containing fungal mats were fixed with 2.5% glutaraldehyde in cacodylate buffer for 2 hr at 4° and then treated with 1% OsO4 in cacodylate buffer for 2 hr at 4°. Samples were slowly dehydrated by first using increasing concentrations of ethanol from 10 to 70% and then using increasing concentrations of acetone from 70 to 100%. Samples were dried in a Balzers CPD 030 Critical Point Dryer and gold-coated. Samples were examined with a JEOL 6460LV Scanning Electron Microscope.
Results
∆gcnE strain does not undergo asexual development
Reyes-Dominguez et al. (2008) noted that deletion of the SAGA/ADA components gcnE or adaB resulted in strongly reduced conidiation. Here, we followed the developmental process in complete medium in a time-course experiment. As shown in Figure 2A, conidiophore heads were already evident after 10 hr of induction in a wild-type strain. After 72 hr of induction, conidiophores were completely mature with heads displaying the regular cylindric morphology. However, the ∆gcnE mutant strain did not show any evidence of conidiophore formation even after 72 hr of induction. Complementation of the ∆gcnE deletion restored the conidiation defects (Figure S1).
Figure 2.
The ∆gcnE mutant is impaired in conidiation. (A) Wild type (WT) and ∆gcnE strains were grown vegetatively for 18 hr, and then conidiation was induced in complete medium. Progression of the developmental program was followed under the stereo microscope at the indicated time points. Conidiophore heads were evident after 10 hr of induction in the wild-type strain. Yellow conidia were evident 24 hr after induction. No such structures were seen in the ∆gcnE strain even after 72 hr of induction. (B) Comparision of the conidiation phenotype of wild-type and ∆gcnE strain with the phenotypes of the mutants in the central regulatory pathway (∆brlA, ∆abaA, or ∆wetA) after 4 days of growth. The brlA mutant produced the stalk cells and then continued growing rather than developing the conidiophore vesicles, metulae, phialides, and conidia. Mutations in abaA and wetA interfered in later stages of conidiophore development and were capable of producing white structures corresponding to the vesicles, metulae, and phialides. The ∆gcnE strain resembles a ∆brlA phenotype. (C) SEM images of the wild-type and ∆gcnE strains grown for 1 week. A very low density of immature conidiophores can be observed in the ∆gcnE strain, compared to the complete development of the wild-type conidiophores. Bar, 50 µm. (D) Details of SEM images comparing the wild-type conidiophores with the aberrant ∆gcnE conidiophore morphologies (indicated by arrows). Arrows indicate details of aberrant conidiophores. The double-line arrow points to a severe example where sterigmata cells seem to bud off from a hyphal or stalk cell. A higher magnification of this example is shown as a separate image at the top right. Bar, 10 µm, except for the top right image where the bar corresponds to 5 µm.
The phenotype of the deletion strain was compared to a set of strains harboring deletions in genes of the central regulatory pathway (brlA, abaA, or wetA) to search for the step at which conidiation was blocked. As shown in Figure 2B, the brlA mutant produced the stalk cells and then continued growing rather than developing the conidiophore vesicles, metulae, phialides, and conidia, which gave it a bristly appearance under the stereo microscope. Mutations in abaA and wetA interfered in later stages of the conidiophore development, and white structures corresponding to the vesicles, metulae, and phialides could be observed under the stereo microscope. The phenotypic differences between abaA and wetA mutants with regards to the formation of conidia could not be observed at this magnification. Nevertheless, the phenotype of ∆gcnE did not resemble ∆abaA or ∆wetA strains, suggesting that developmental defects probably originate in genes upstream of abaA. Indeed, the ∆gcnE mutant looked more like a ∆brlA strain. When the ∆gcnE strain was allowed to grow for 1 week, some conidiophores could be observed (Figure 2C). The colony showed a very low density of conidiphores as compared to a wild-type strain. In addition, higher SEM magnifications revealed that the conidiophores were not completely developed, harboring rows of four conidia at most, even after 1 week of growth. Most remarkably, these conidiophores displayed aberrant morphologies, for example, a conidium arising from a hyphal tip, rama growing out of stalk cells, or sterimagta cells budding off what could be stalk or hyphal cells (Figure 2D).
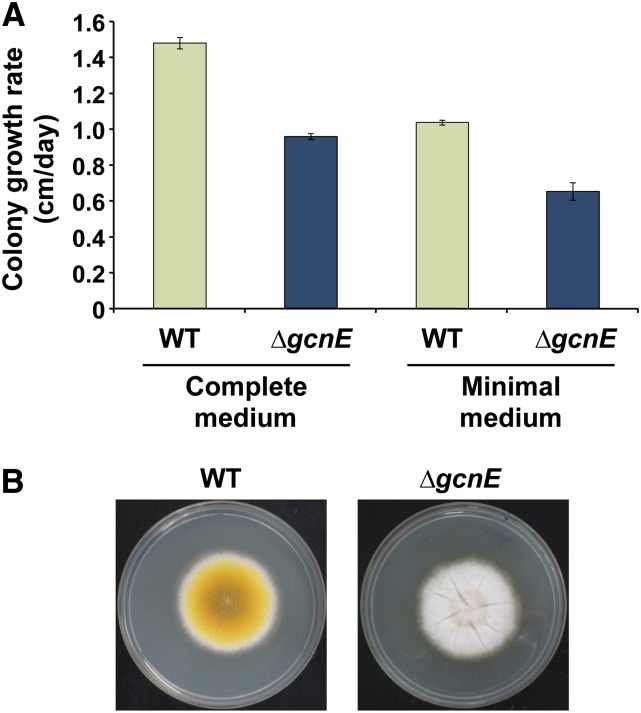
We reasoned that the conidiation defects could be due to the growth reduction previously reported (Reyes-Dominguez et al. 2008). To test this, we quantified the linear growth rate of both the wild-type and an isogenic ∆gcnE mutant strain on complete and minimal solid media (Figure 3A). Consistent with the previous report, the wild-type strain grew faster than the ∆gcnE strain on both complete and minimal media. However, the growth reduction observed in the mutant strain was not strong enough to explain the conidiation defects. When both the wild-type and the ∆gcnE strains were point-inoculated on plates and allowed to grow until they reached the same colony size (diameter), the wild type showed strong conidia developement whereas the mutant strain did not show any signs of conidiation (Figure 3B). This suggests that the growth reduction in the mutant strain is not responsible for the conidiation defects.
Figure 3.
Differences in growth rate do not explain the conidiation defects in ∆gcnE. (A) Growth of wild-type and ∆gcnE strains was followed on complete and minimal solid media over a period of 5 days. The linear growth rate of the mutant was only slightly lower in comparison with the wild type on both media. The growth rate is shown as the increment in the colony diameter on solid media per day. Error bars show the standard error of at least three independent experiments performed in duplicates. (B) Wild type and ∆gcnE strains were point-inoculated on complete media plates and allowed to grow at 37 °C. Plates were photographed after the colonies reached the same size.
Transcriptome analysis of conidiation
The fact that the gcnE-deletion mutant phenotype was most similar to the ∆brlA mutant suggested that conidiation is blocked at an early stage of development, and no or very few conidiophore heads are produced. Thus, the mutant cells are most likely defective in the expression of the upstream developmental regulators or genes in the central regulatory pathway. As chromatin modifyers impact on the expression of a large set of genes, we globally compared gene expression in the wild type and in the ∆gcnE during vegetative growth and development. In a first approach, the transcriptomes of wild-type cells grown vegetatively in liquid medium (time 0, non-induced) and at 10 hr after induction of conidiation on solid medium (time 10 hr, inducing conditions) were compared using two colors expression microarray. At this time, genes in the central regulatory pathway are already activated. The analysis of the data by Empirical Bayes Test statistics revealed that 1225 genes were differentially regulated (i.e., 13.6% of the total number of genes in the chip) between these two conditions. Of these 1225 differentially regulated genes, 600 were downregulated and 625 were upregulated (Figure S2; Table S3 and Table S4 list the Top 25 up- and downregulated genes). This suggests that after 10 hr of induction a major reprogramming of the gene expression profile occurred in the conidiating cultures. Some of the upregulated genes are relevant for the regulation of conidiation (see Table 1 for a list of genes), for example, the fluffy gene flbC, and the central regulatory cascade of transcriptional activators (brlA, abaA, and wetA) involved in the temporal and spatial regulation of the conidiation genes (Mirabito et al. 1989). Other genes involved in conidiation were also found to be upregulated, such as vosA, medA, ivoB, yA, rodA, and dewA. Gene ontology (GO) term analysis of the significant genes upregulated during conidiation revealed that some terms were enriched (see Figure S3), e.g., carbohydrate metabolism, in which 60 genes of the 584 upregulated genes having GO descriptions were induced, including polysaccharide (25), pectin (7), alcohol (31), xylan (6), and pentose (9) metabolism genes. Another GO term was found to be involved in secondary metabolism and toxin synthesis corresponding to 20 genes of the 584 upregulated genes having GO descriptions (e.g., 8 genes of the sterigmatocystin biosynthesis cluster and the regulator aflR). Another GO group that was enriched is related to cell-wall biogenesis (18 of 584), which includes conidiation genes involved in the synthesis of the spores layers (dewA, rodA, wetA). sdeA and sdeB, which are required for development in A. nidulans (Wilson et al. 2004), belong to another enriched GO.
Table 1. Differentially regulated genes involved in development (sexual or asexual) and secondary metabolism in A. nidulans.
| WT vegetative vs. conidiation | ∆gcnE vegetative vs. conidiation | |||||
|---|---|---|---|---|---|---|
| Gene name | Gene code | qmediana | Log qmedian | qmedian | Log qmedian | Description of gene function |
| dewA | AN8006 | 160.8 | 5.1 | — | — | Hydrophobin, protein of the conidium wall responsible for hydrophobicity of conidium surface |
| yA | AN6635 | 28.2 | 3.3 | — | — | Conidial laccase (p-diphenol oxidase) involved in dark-green pigment production of conidium wall |
| pclA | AN0453 | 11.6 | 2.4 | 5.4 | 1.7 | G1/S cyclin: mutants produce abnormal conidiophores with extra layers of phialides. |
| ivoB | AN0231 | 11.1 | 2.4 | 3.7 | 1.3 | Conidiophore-specific phenol oxidase |
| wetA | AN1937 | 11.0 | 2.4 | — | — | Regulatory protein involved in conidial development |
| aflR | AN7820 | 10.0 | 2.3 | — | — | Transcriptional activator of the sterigmatocystin biosynthesis gene cluster |
| abaA | AN0422 | 8.9 | 2.2 | — | — | TEA/ATTS domain transcriptional activator involved in regulation of conidiation; required for phialide differentiation. |
| brlA | AN0973 | 5.9 | 1.8 | — | — | C2H2 zinc-finger transcription factor, master regulator of conidiophore development |
| rodA | AN8803 | 5.5 | 1.7 | — | — | Hydrophobin |
| medA | AN6230 | 5.3 | 1.7 | — | — | Protein involved in regulation of conidiophore development; required for normal temporal expression of brlA. |
| hogA | AN1017 | 4.2 | 1.4 | — | — | MAPK involved in osmotic stress response; required for sexual development and conidiation. |
| flbC | AN2421 | 3.7 | 1.3 | — | — | C2H2 zinc-finger transcription factor; involved in regulation of conidiophore development. |
| vosA | AN1959 | 3.6 | 1.3 | — | — | Nuclear protein involved in spore formation and trehalose accumulation |
| imeB | AN6243 | 0.4 | −0.9 | — | — | Serine/threonine protein kinase involved in light-mediated regulation of sexual development and sterigmatocystin production |
| nsdC | AN4263 | 0.3 | −1.2 | — | — | C2H2 zinc-finger transcription factor; required for sexual development. |
| laeA | AN0807 | 0.2 | −1.9 | — | — | Methyltransferase-domain protein: velvet complex component composed of VelB, VeA and LaeA; coordinates asexual development in response to light; regulates secondary metabolism; and is required for Hülle cell formation |
| pipA | AN2513 | 0.1 | −1.9 | — | — | Serine/threonine protein kinase involved in hyphal growth and asexual development |
| sskA | AN7697 | — | — | 0.2 | −1.6 | Response regulator, part of a two-component signal transducer involved in the HOG-signaling pathway that regulates osmotic stress response; null spores are heat labile and lose viability at 4° |
| rosA | AN5170 | — | — | 13.6 | 2.6 | Zn(II)2Cys6 transcription factor; negative regulator of sexual development |
—, not differentially regulated under those conditions in that strain.
qmedian values >1 (or positive log qmedian values) indicate that the gene is expressed during conidiation, while qmedian values <1 (or negative log qmedian values) indicate that the gene is expressed during vegetative growth. An absence of value indicates that gene is not differentially expressed in that strain.
GO term analysis of the significant genes downregulated during conidiation (i.e., vegetative genes) identified small molecule metabolism (89 genes of the 579 downregulated genes having GO descriptions), which included metabolism of ketone (52), carboxilic acids (51), and cellular nitrogen (39) and biosynthesis of heterocycle (cofactor and coenzyme) (25) and nucleotides (17) (see Figure S3). This suggests that genes involved in primary metabolism were expressed more strongly during vegetative growth, while genes related to development or secondary metabolite production were expressed at a higher level during the conidiation program.
Transcriptome analysis of gcnE-dependent and independent control of gene expression during development
In contrast to the 1225 genes differentially regulated in the wild-type strain, only 319 genes were differentially regulated in the ∆gcnE strain when vegetative and conidiation conditions were compared (Figure S2). This corresponds to only 26% of the number of genes (319 in the ∆gcnE strain vs. 1225 in the wild type) differentially regulated in the wild-type strain. Therefore, deletion of gcnE appeared to affect the expression of a large number of genes regulated during development. Of these differentially expressed genes, 181 genes were upregulated and 138 genes were downregulated in the ∆gcnE mutant (Figure S2; Table S5 and Table S6 list the Top 25 up- and downregulated genes). In agreement with the phenotype of the mutant, some regulators of conidiation were not upregulated during conidiation in the ∆gcnE strain, for example, brlA, abaA, wetA, vosA, and medA (Table 1). In addition, genes required for the synthesis of secondary metabolites (such as laeA or the sterigmatocystin gene cluster) were not upregulated either.
Several GO groups mainly related to primary metabolism showed a higher expression level during vegetative growth in the mutant, but only one GO group, namely xylan metabolism, was more strongly expressed in the gcnE mutant under conidiation conditions (Figure S4). Therefore, wild-type and ∆gcnE strains shared most of the GO terms of genes upregulated during vegetative growth but not during conidiation, suggesting that deletion of gcnE affected mainly the regulation of genes during development. Of the 625 genes upregulated during development in the wild type, 41 were also upregulated in the ∆gcnE, which suggests that these genes are gcnE-independent. Seventy-four genes appeared to be gcnE-independent in the downregulated genes (Figure S2).
Transcriptome analysis reveals that GcnE is involved in the regulation of conidiation and secondary metabolism genes
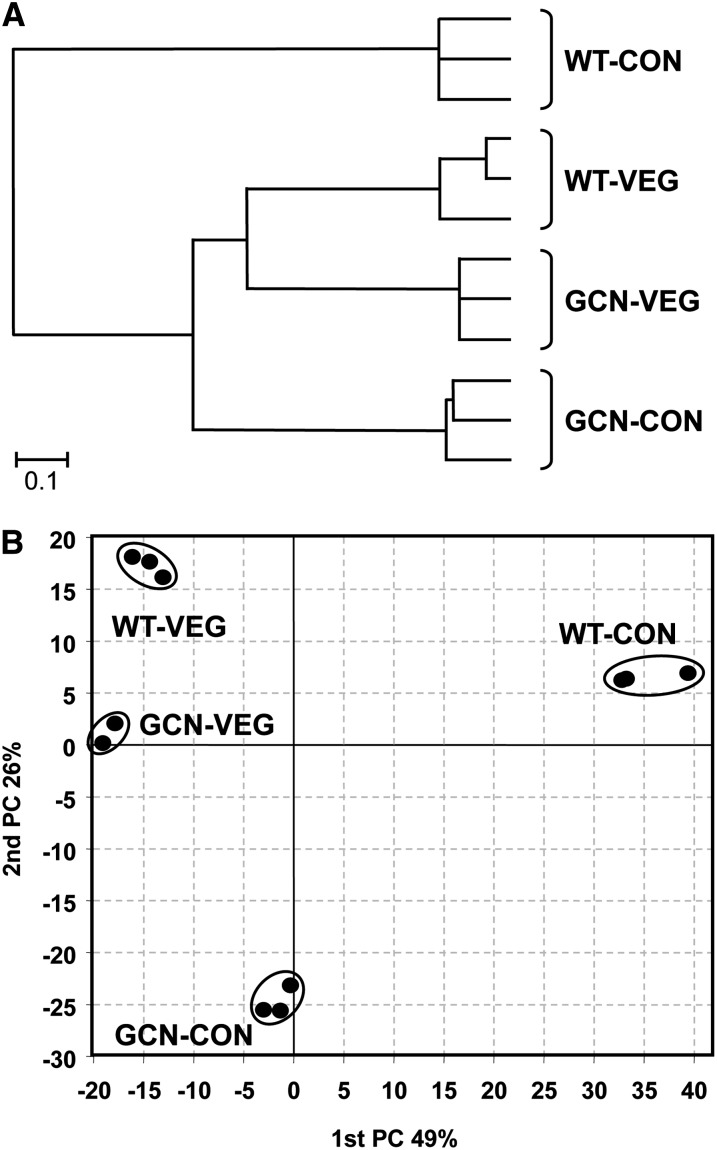
The transcriptome data of the wild-type and ∆gcnE cells was further analyzed by ANOVA to allow the comparision of all four conditions at the same time, i.e., wild-type vegetative, wild-type conidiation, ∆gcnE vegetative, and ∆gcnE conidiation. Using this type of analysis, 1162 genes were found to be differentially expressed in at least one of the conditions. This corresponds to 10.9% of the total number of predicted A. nidulans genes. The expression pattern of these 1162 differentially expressed genes was grouped by using centroid linkage clustering of the euclidean distances. The resulting dendrogram shows that the ∆gcnE strain grown under conidiation conditions clustered together with vegetative cultures of both the wild-type and the mutant strain (Figure 4A). We further analyzed the differentially expressed gene set by PC analysis to assess the contribution of the genetic background or the growth mode to the gene expression pattern. PC analysis assigns coordinates (components) to the variation in gene expression, representing the largest, second largest, third largest, and so on variance in the corresponding axis. As shown in Figure 4B, the major variation between vegetative growth and conidiation in the wild-type strain was depicted in the first PC (the x-axis), while the second PC (the y-axis) showed small differences between these two conditions in comparison (∼50 and 11 units, respectively). On the other hand, differences between the wild-type strain and the ∆gcnE mutant appeared mainly in the second PC (y-axis) and not in the x-axis, when the strains were grown vegetatively (∼4 units in the x-axis vs. 16 units in the y-axis). This difference between the wild type and mutant strains in the y-axis strongly increased when conidation was induced (∼31 units). PC analysis assigned 49% of the variation to the mode of growth (vegetative vs. conidiation) and 26% to the genetic variation (wild type vs. ∆gcnE). These results mean that the gene expression profile of the vegetative wild-type strain was similar to the ∆gcnE strain under both vegetative and conidiation conditions in the x-axis/first PC (∼4 and 13 units, respectively; Figure 4B). In other words, the gcnE mutant under conidiation conditions was more similar to the wild-type strain growing vegetatively than conidiating (the gene expression profile of the gcnE mutant was similar to the vegetatively growing wild type regardless of the growth mode of the mutant). The results of this statistical analysis enforces the view that GcnE plays a more important role in regulation of development and seems to be less involved in the regulation of transcription under the conditions of vegetative growth used in this set of experiments. Among the top 20 genes differentially expressed during conidiation in the wild-type strain (positive side of the first PC), 9 genes are known to have a role in conidiation or secondary metabolism (Table S7). The negative side of the first PC includes genes involved in oxidoreduction or other metabolic activities, such as hydrolases and peptidases (Table S8). Using this approach, we thus identified a group of genes that were specifically expressed in the wild-type strain during conidiation but were not expressed in the gcnE mutant under any condition.
Figure 4.
Global expression analysis of wild-type and ∆gcnE strains growing under vegetative or conidiation conditions. Both strains were grown vegetatively for 18 hr, and then conidiation was induced for 10 hr. The global expression of genes under the four conditions (wild-type vegetative, WT-VEG; wild type-conidiation, WT-CON; ∆gcnE vegetative, GCN-VEG; ∆gcnE conidiation, GCN-CON) was compared by using microarray hibridization. A total of 1162 differentially expressed genes were identified by ANOVA. (A) A dendogram was obtained by centroid linkage clustering using euclidean distances of the 1162 differentially regulated genes in the 12 samples (four conditions with three biological replicates each). The ∆gcnE strain grown under conidiation conditions was more similar to vegetative growth than to the conidiating wild type. (B) PC analysis of the genes differentially regulated under at least one of the four different conditions. The x-axis shows the first PC with a variation of 49% due to the growth mode (vegetative vs. conidiation). The y-axis shows the second PC with a variation of 26% due to the genetic background (wild type vs ∆gcnE). The results obtained by clustering (A) and PC (B) analysis are in agreement.
Two interesting genes appearing in this list were nkuA and nkuB. Knockout strains of nkuA have become widely used in Aspergillus and Neurospora laboratories after the discovery that deletion of the KU80 or KU70 homologs results in a high rate of homologous integration but does not affect development (Ninomiya et al. 2004; da Silva Ferreira et al. 2006; Krappmann et al. 2006; Nayak et al. 2006). However, both genes were found to be upregulated during conidiation, and their induction seems to be GcnE-independent (upregulated also in the ∆gcnE mutant). A more complete analysis of the GO terms of genes found to be differentially regulated by ANOVA is shown in Figure S5, Figure S6, Table S7, Table S8, Table S9, and Table S10.
brlA is a major target of GcnE
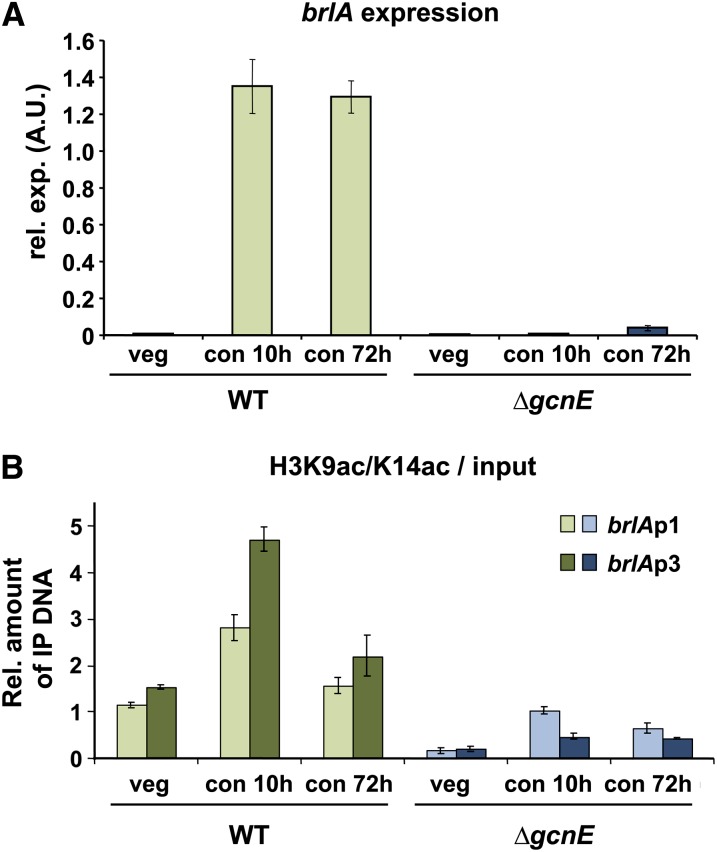
Analysis of the transcriptomes revealed that conidiation genes and their regulators were induced in the wild-type strain after 10 hr of induction, but most of these genes remained unchanged in the ∆gcnE strain (Table 1). As expected, we found brlA as an induced gene in the wild type, but this central regulator was not upregulated in the ∆gcnE strain. To get further details and to confirm transcriptome data, we studied the expression of brlA by RT-qPCR in the wild-type and the gcnE mutant strain grown under the same conditions as employed for the microarray experiment and, in addition, in cultures harvested 72 hr after induction of conidiation (Figure 5A). In the wild-type strain, brlA expression was high at both time points whereas the ∆gcnE strain did not show any accumulation of brlA mRNA after 10 hr. Interestingly, brlA mRNA can be detected in this strain at 72 hr post-induction (still ∼30-fold lower than in the wild-type strain), which is in agreement with microarray data and with the ΔgcnE phenotype, in which some conidia are produced upon prolongued incubation on solid media.
Figure 5.
brlA is not expressed and acetylation of histone H3K9/K14 at the brlA promoter is reduced in the ∆gcnE strain. (A) Both wild-type and ∆gcnE strains were grown vegetatively for 18 hr, and then conidiation was induced for 10 or 72 hr. RNA was isolated and gene expression was quantified by RT-qPCR. Data are shown normalized to the tubulin gene (benA) as an internal standard. (B) ChIP was carried out by immunoprecipitation of cross-linked DNA with an antibody recognizing acetylated histone H3K9ac and H3K14ac, followed by qPCR analysis of the promoter regions. brlA showed an increase in the immunoprecipitated DNA in both distal (brlAp1) and proximal (brlAp3) regions of the promoter in the wild type. In the ∆gcnE strain, acetylation levels were grossly reduced and conidiation-specific increases were not observed. Values were normalized to input DNA (before immunoprecipitation) and are shown as the mean with standard errors of the mean of at least three biologically independent experiments.
To find out whether the effect of gcnE deletion on brlA is direct or indirect, we determined the acetylation pattern of histone H3K9 and H3K14 at the brlA promoter by ChIP. Because the promoter of brlA covers >2 kb from the ATG of brlAα (Garzia et al. 2010; Kwon et al. 2010), we employed two different primer pairs. Primers brlAp1 were located at a distal position from the ATG of brlAα (–2303 to −2483 bp), spanning the FlbB-binding site, while primers brlAp3 were located at a proximal position to the ATG (−60 to −245 bps). The acetylation levels of H3K9 and H3K14 increased after induction of conidiation in both regions of the promoter in the wild-type strain (Figure 5B). The levels of acetylation were higher at 10 than at 72 hr after induction of conidiation. The overall acetylation pattern was similar in both regions of the promoter although the levels were higher in the proximal region to the ATG than in the distal region. This can be explained by the fact that highly acetylated nucleosomes +1 in the open reading frames are not evicted during transcriptional activation (in contrast to promoter nucleosomes) (Workman 2006), and the DNA fragments encompassing nucleosome +1 are captured by the proximal PCR primers. On the contrary, in the ∆gcnE strain the acetylation levels at the promoter of brlA were lower than in the wild-type strain and did not increase over the basal levels of the wild-type strain growing vegetatively. This was consistent with the lack of brlA expression and, consequently, with the absence of conidiophore formation in the mutant. Analysis of the total amount of histone H3 at the brlA promoter by ChIP revealed that the total amount of histone H3 decreased in the wild type after induction of conidiation and reached a minimum at 72 hr (Figure S7). In contrast to the wild-type strain, the total levels of histone H3 did not decrease, but even increased in the ∆gcnE strain upon induction of conidiation, consistent with an inactive promoter (Figure S7).
Inhibitors of histone deacetylation do not recover conidiation in ΔgcnE
Surprisingly, although the histone H3K9 and H3K14 acetylation levels were below the wild-type basal levels in the ∆gcnE strain, there was a slight increase in acetylation after the induction of conidiation. We reasoned that alternative histone H3 acetyltransferases may be operating at these genes under induction conditions as ∼40 putative acetyltransferases are present in the genome of A. nidulans (Nützmann et al. 2011). To test this possibility, we employed HDAC inhibitors to block deacetylation. This presumably would lead to increased acetylation levels of histone H3 and may recover conidiation in the ∆gcnE mutant. Trichostatin A was already shown to be an effective HDAC inhibitor in A. nidulans in previous studies (Shwab et al. 2007). Addition of trichostatin A or a cocktail of inhibitors (trichostatin A + butyric acid + valproate) did not result in restoration of conidiation, not even partially (Figure S8). Therefore, the most plausible explanation is that the histone H3 acetylation levels in the ∆gcnE strain corresponded to background levels and that a functional SAGA complex is necessary for brlA expression.
Expression of the upstream regulatory genes controlling conidiation in the ∆gcnE strain is deregulated
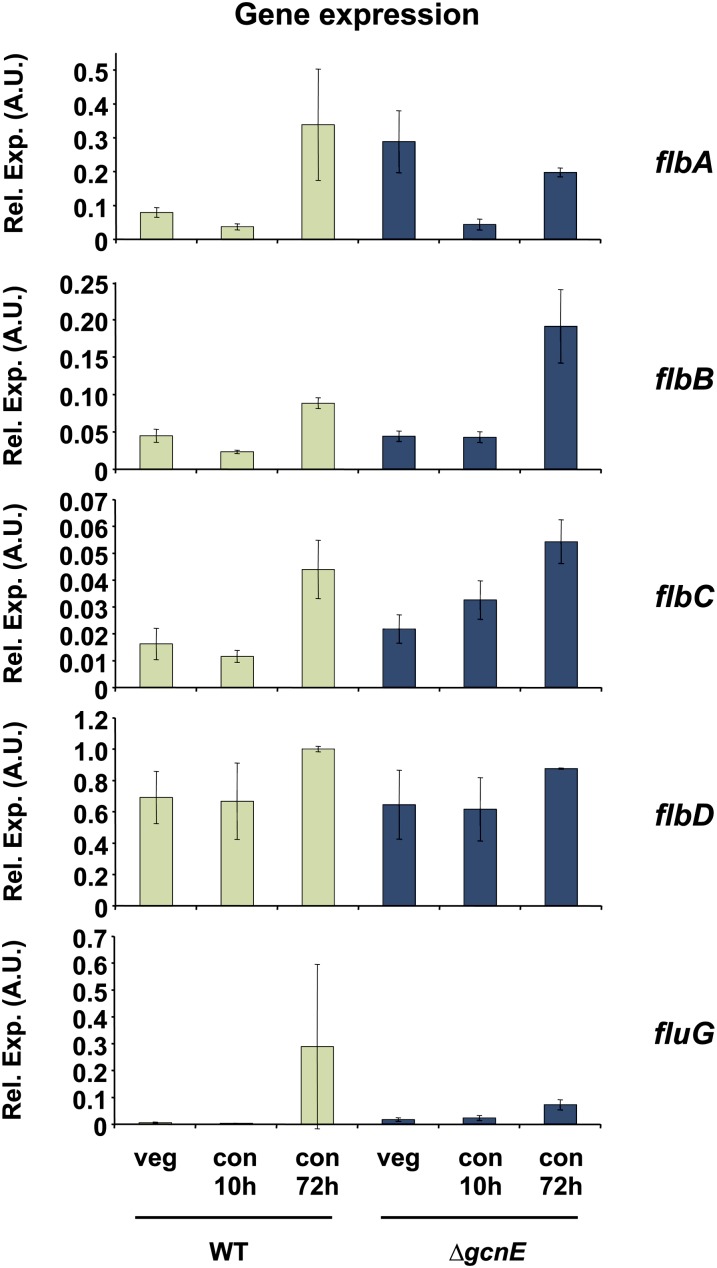
There are three parallel routes for the activation of conidiation, consisting of FlbA, FlbB/D, and FlbC (Adams et al. 1998; Etxebeste et al. 2010). The transcriptome studies revealed a slight upregulation of flbC, one of the upstream factors for conidiation genes, and that this pattern was affected by the gcnE deletion (Table 1). To get further details and to confirm transcriptome data, we tested the expression of flbC and four of the other upstream regulators in the wild-type and gcnE mutant strain grown under the conditions described above in Figure 5. As shown in Figure 6, none of these genes were upregulated in the wild-type strain after 10 hr of induction (the conditions used for the microarray experiment). However, after a longer period of induction (72 hr), four of these genes (flbA, flbB, flbC, and flbD) showed higher steady-state mRNA levels compared to vegetative mycelia or after 10 hr of induction in the wild-type strain. When mRNA levels of these five genes were compared between both strains in vegetative mycelia, flbA showed a threefold higher expression in the ∆gcnE strain than in the wild type, whereas the other genes were basically identical. There was no significant difference between wild type and ∆gcnE in expression of the upstream regulators at time point 10 hr of induction with the exception of flbC, but interestingly, at 72 hr, flbA expression is reduced whereas flbB expression is around twofold higher in the gcnE mutant compared to the wild-type strain. In the case of fluG, there was a big variation from sample to sample at the 72-hr time point, so no conclusion could be drawn from these results. These slight differences in mRNA levels suggest that the lack of conidiation in ∆gcnE probably did not originate from defects in fluffy gene expression. This is also consistent with the fact that expression of the fluffy genes was already detected in vegetative mycelia and that their transcriptional upregulation is not the critical regulatory point for their function in the transcriptional activation of brlA.
Figure 6.
Expression of the fluffy genes during conidiation is deregulated in the ∆gcnE strain. Both wild-type and ∆gcnE strains were grown vegetatively for 18 hr, and then conidiation was induced for 10 or 72 hr. RNA was isolated and gene expression was quantified by RT-qPCR. Data are shown normalized to the tubulin gene (benA) as an internal standard. Values are the mean and standard error of the mean of at least three independent experiments.
Consistent with the moderate transcriptional activation of flbA, flbB, flbC, and flbD genes, their promoters showed higher levels of H3 acetylation during induction of conidiation (data not shown). This increase was associated with a decrease in the amount of total histone H3 present at these promoters probably due to partial eviction of nucleosomes from these regions. This difference was not observed in the mutant strain that showed very low H3 occupancy at all fluffy gene promoters before and after conidial induction (data not shown).
Are there additional targets mediating the GcnE effects on development?
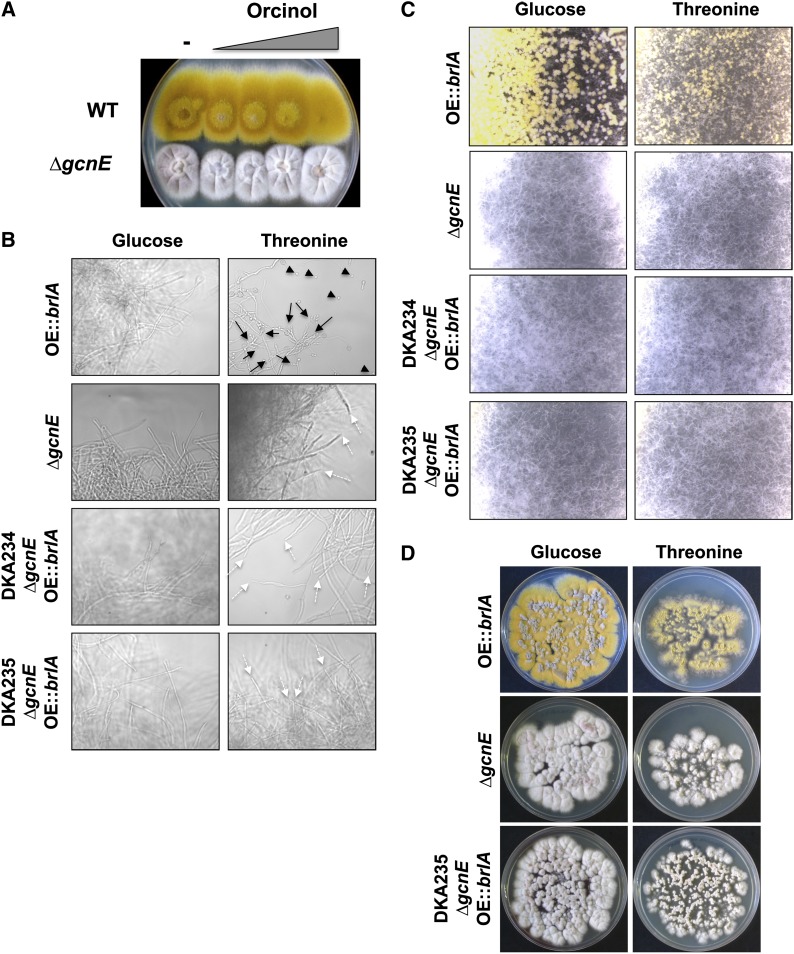
During the analysis of the microarray data, we observed that the orcinol/orsellinic acid cluster was not expressed in the ∆gcnE strain. This is in agreement with a previous report by Nützmann et al. (2011). Diorcinol is a derivative of orsellinic acid and functions together with dehydroaustinol, one of the signals required for the induction of conidiation (Rodriguez-Urra et al. 2012). One possibility is that the absence of the expression of this cluster contributes to the conidiation defects. However, addition of different concentrations of orcinol ranging from 50 µg to 50 mg did not recover the conidiation defects of the ∆gcnE strain (Figure 7A). Furthermore, the deletion of the polyketide synthase orsA, responsible for the biosynthesis of orsellinic acid (Schroeckh et al. 2009), a precurssor of diorcinol, did not show any conidiation defects (data not shown).
Figure 7.
GcnE has additional as-yet-unidentified targets mediating the developmental effects. (A) Wild-type and ∆gcnE strains were pregrown for 24 hr before addition of different concentrations of orcinol (50 µg to 50 mg) on top of the colony. Plates were incubated for 3 additional days and photographed. The highest concentration of orcinol had some slightly negative effects on colony development in both strains. (B) Strains indicated at the left were pregrown for 24 hr in liquid media under repressing conditions (glucose) and then transferred to fresh liquid medium containing inducing threonine or repressing glucose, and incubation was continued for an additional 24 hr. Fungal pellets were photographed under the light microscope. The parental strains harbored either a construct overexpressing brlA from the alcA promoter (OE::brlA) or the gcnE deletion (∆gcnE). Two independent strains of the cross progeny (DKA234, DKA235) were used in this experiment. Black arrows indicate conidiophore-like structures, black arrowheads point to individual conidia produced in liquid cultures, and white dotted arrows point to vegetative hyphal tips. (C) Strains were pregrown as in B for 24 hr under repressing condintions but then transferred to solid medium containing threonine or glucose, and incubation was continued for 1 day. Fungal colonies were photographed under a stereo microscope at the same magnification. The OEbrlA strain (brlA+; alcA(p)::brlA) conidiated on glucose plates due to brlA expression from its native promoter. Two independent strains of the progeny were also used in this experiment. (D) Strains pregrown for 24 hr under repressing condintions (as in B) were transferred to solid medium containing threonine or glucose, and incubation was continued. Plates were photographed after 3 days of growth. Growth inhibition could be observed in the strains overexpressing brlA in both the wild-type and ∆gcnE background only under brlA-inducing conditions (threonine).
Therefore, this result further implied that the conidiation defects of the ∆gcnE strain could be directly related to SAGA function at the brlA promoter. We tested this by expressing the brlA gene under the control of the heterologous inducible promoter of the alcA gene at an ectopic location (Adams et al. 1988). ∆gcnE alcA(p)::brlA strains were constructed by crossing. The parentals and two strains from the progeny were grown in liquid medium for 24 hr and then transferred to inducing (threonine) or repressing (glucose) liquid medium and grown for another 24 hr. Inspection of the fungal pellets by light microscopy (Figure 7B) revealed that the wild-type strain harboring the alcA(p)::brlA construct produced primitive conidiophores and conidia under inducing conditions as previously reported (Adams et al. 1988). However, none of the ∆gcnE alcA(p)::brlA strains produced any of these primitive conidiophores or conidia. The experiment was repeated twice with two independent strains from the progeny. Next, we transferred the strains grown for 24 hr in glucose liquid media (repressing conditions) to solid media containing threonine (inducing) or glucose (repressing) conditions. The ∆gcnE alcA(p)::brlA could not conidiate even under these conditions (Figure 7C). However, it is interesting that growth restriction upon induction of alcA(p)::brlA on threonine was observed in both the wild-type and the ∆gcnE strains overexpressing brlA (Figure 7D). This effect was already observed by Adams et al. (1988) and may be suggestive of a BrlA role in halting vegetative growth, perhaps through crosstalk with the FlbA/G-protein-signaling pathway regulating vegetative growth.
Discussion
The data obtained during this work revealed that GcnE is the only nonessential histone modifier found so far with an essential function in fungal development. In previous work, we and others have established that histone acetylation plays only a minor role in the regulation of some selected primary metabolic systems (Reyes-Dominguez et al. 2008; Georgakopoulos et al. 2012) but significantly regulates secondary metabolism (Shwab et al. 2007; Nützmann et al. 2011, 2013; Bok et al. 2013). Data presented here enlarge our picture of GcnE function to a genome-wide scale, and from these experiments it is becoming clear that GcnE is a minor regulator of primary metabolism but an essential component in driving A. nidulans developmental processes. In the diverse set of reversible histone modifications, acetylation and methylation have been the most extensively studied ones in filamentous fungi (Gacek and Strauss 2012). It has been shown that the histone H3K9 methyltransferase ClrD and the heterochromatin-protein 1 (HepA) are regulators of secondary metabolite gene clusters (Reyes-Dominguez et al. 2010; Gacek and Strauss 2012). Deletion of the histone H3K9 methyltransferase clrD or the histone H3K4 methyltransferase cclA in A. nidulans has no growth or conidiation phenotype (Bok et al. 2009; Reyes-Dominguez et al. 2010). However, a decrease in radial growth and delayed conidiation due to later brlA expression is observed in the equivalent A. fumigatus clrD deletion mutant (Palmer et al. 2008). The histone deacetylase RpdA is essential for growth in A. nidulans, and, consequently, a direct and unequivocal effect on development has not been tested yet (Tribus et al. 2010). The histone H4K12 acetyltransferase EsaA is also essential for growth and cooperates with the general secondary metabolite regulator LaeA to mediate histone H4 acetylation and transcriptional activation of selected secondary metabolite clusters (Soukup et al. 2012), but due to its essential nature the involvement in conidiation remains elusive. Similarly, deletion of the histone deacetylase hdaA was reported to have effects on secondary metabolism but not on development (Shwab et al. 2007; Bok et al. 2009). Therefore, other histone modifiers either have shown a minor role during development or are essential for growth, while GcnE is required for conidiation but not essential for growth. The SAGA complex is also involved in the regulation of development in higher eukaryotes, such as plants (Servet et al. 2010), and metazoans (Spedale et al. 2012). In Arabidopsis, AtGCN5 plays an essential role in the development of root and shoot and flower meristems, leaf-cell differentiation, and responses to light. AtGCN5 also appears to regulate the expression of a large number of genes, likely mediated by direct or indirect interactions with DNA-binding transcription factors (Servet et al. 2010). In metazoans, it was suggested that Gcn5 may be required to maintain pluripotent states and is important for the differentiation of rat mesenchymal stem cells into cardiomyocytes. Indeed, loss of Gcn5 resulted in a hard-pack chromatin structure at the cardiomyocyte-specific genes GATA4 and NKx2.5 and elevated levels of apoptosis during embrionic development (Lin et al. 2007; Li et al. 2010). It is intriguing that, while metazoans have evolved four HAT complexes acetylating histone H3 specialized in different cellular processes (Spedale et al. 2012), A. nidulans has only one.
The SAGA complex plays a general role in transcriptional activation in yeasts. TAFII145 and Gcn5 are apparently functionally redudant in yeast (Lee et al. 2000), although there is some specialization of the SAGA complex in stress-related genes (Huisinga and Pugh 2004). Although the SAGA complex is very similar in A. nidulans to the yeast counterpart, the role of GcnE seems to be significantly different from the role of its orthologs in yeasts. Thus, according to genome-wide expression analysis and the observation of mutant phenotypes, it appears that the main role of GcnE is to regulate development and some specific secondary metabolism gene clusters in A. nidulans. Some of the secondary metabolites can be considered as “weapons” utilized only under stressing conditions in nature as a defense mechanism. For example, it was found that GcnE played a major role during the induction of biosynthetic gene clusters of sterigmatocystin, terrequinone, and penicillin (Nützmann et al. 2011). An interesting case is polyketide orsellinic acid, which is produced by A. nidulans in response to the interaction with a streptomycete species in a GcnE-dependent manner (Nützmann et al. 2011). Synthesis of orsellinic acid derivatives F9775A and -B, which is induced in a ∆veA strain, is lost in the double-mutant ∆gcnE ∆veA (Bok et al. 2013). Thus, the reported role of GcnE in the regulation of secondary metabolism is consistent with our microarray expression analysis. Orsellinic acid is also the precursor of diorcinol, which makes an adduct with the bioactive compound dehydroaustinol, produced by FluG, to induce conidiation (Rodriguez-Urra et al. 2012). Although the absence of GcnE activity could lead to a lack of this conidiation inducer adduct, addition of external orcinol did not restore conidiation. In support of this, the deletion of the polyketide synthase orsA responsible for the biosynthesis of orsellinic acid (Schroeckh et al. 2009) did not show any conidiation defects either. Therefore, a scenario in which brlA expression is not turned on in the gcnE mutant after induction due to the lack of the inducer adduct seems unlikely. Instead, GcnE appears to be responsible for histone H3K9/K14 acetylation at the brlA promoter, which in turn is a prerequisite for brlA expression. Consequently, deletion of gcnE blunts brlA expression under inducing conditions. However, this is only a part of the whole picture because forced expression of brlA from the inducible alcA promoter could not restore conidiation in a ∆gcnE background. As expression of upstream regulators of the fluffy family of genes (flbA, flbB, flbC, flbD, and fluG) was also not significantly affected by gcnE deletion and the phenotype does not conform to the mutants of the central regulatory pathway abaA-wetA either, we have to assume that other regulators of conidiation may be “hidden” targets of this SAGA complex component. One such target may be the velvet complex members veA, velB, or velC (Bayram et al. 2008b) or the light receptors fphA, lreA/B, or cryA (Bayram et al. 2008a; Purschwitz et al. 2008). However, we did not observe any significant change in expression of these regulators and photoreceptors comparing the wild type and the gcnE mutant, and these genes are not even responsive to induction of conidiation in the wild type. Thus it is unlikely that some of these known genes involved in developmental regulation are targets of the SAGA complex. At the moment it remains elusive which of the differentially regulated genes (apart form brlA) may be responsible for the strong conidiation-deficient phenotype of the gcnE deletion strain. In addition, the SAGA complex participates in more cellular functions as it is necessary not only for the activation of gene expression but also for transcriptional elongation, splicing, nuclear mRNA export, and as a general platform for the recruitment of regulatory factors (Millar and Grunstein 2006; Baker and Grant 2007; Gunderson and Johnson 2009; Gunderson et al. 2011). Therefore, the conidiation defects in the ∆gcnE strain could be mediated through a combination of several targets and diverse molecular activities, which deserve further investigation.
In this study, we compared vegetative cells grown in liquid cultures immediately before and after 10 hr of shift to conidiation conditions and found 1225 genes differentially regulated, of which 625 were upregulated. Garzia et al. (2013) analyzed the conidiation-specific transcriptome after 5 hr of induction and found 2222 genes differentially regulated (corresponding to 20.3% of the genes present in the genome), of which only 187 were upregulated. These numbers are much higher than the 533 genes found to be differentially regulated in response to light in A. nidulans (Ruger-Herreros et al. 2011). It suggests that induction of asexual development actually results in a major cellular reprogramming over time. For example, master regulators of carbon (creA) and nitrogen (areA) metabolism did not appear to be differentially regulated at 10 hr after the induction of conidiation in our analysis, but they were downregulated after 5 hr. The comparison of both transcriptomic experiments at different time points suggests that in a first stage there is a major downregulation of genes expressed in the vegetative phase to produce a growth arrest. In a next stage, many genes are upregulated to accommodate all morphogenetic requirements for asexual reproduction. Ten hours after the transition from vegetative growth to conidiation, the number of upregulated genes approximates to the prediction of 1200 unique mRNAs postulated by Timberlake (1980). However, Martinelli and Clutterbuck (1971) estimated that only between 45 and 150 genes are specifically required for conidiation. This difference could be explained with genes that are not specifically required for conidiation, but rather play additional roles or simply indirectly respond to the changing environmental conditions (exposure to oxygen, light, solid interphase, different nutrient signaling, etc.). For example, the osmotic stress MAPK hogA is still upregulated after 10 hr (as it is after 5 hr) of induction of conidiation, but some of its targets or downstream regulators are not (atfA, srrA, tcsA). None of the chromatin regulators known in A. nidulans appear to be regulated at the transcriptional level upon induction of conidiation (gcnE, adaB, clrD, hepA, cclA, rpdA, dmtA, and laeA), and they do not require GcnE for their constitutive expression (they are not affected by the ∆gcnE deletion). Notably, one of the most heavily affected GO categories found in the list of genes upregulated in the ∆gcnE mutant during conidiation was related to responses to stress and, in particular, to DNA damage (nkuA, nkuB, uvsC, and other putative genes). It can be argued that GcnE is required for genome stability maintenance and/or DNA repair during conidiation, and, consequently, the absence of GcnE may generate DNA damage stress. The function of the spores is not only dispersion in the environment but also protection of the genome (Park and Yu 2012). Indeed, isolation of yeast mutants affected in components of the SAGA/ADA complex showed a phenotype of increased Rad52 foci and sister-chromatid recombination (Munoz-Galvan et al. 2013). Whether GcnE and the SAGA complex play similar roles in filamentous fungi is not known yet, but the importance of GcnE in conidiospore production may justify speculations of a similar role in these organisms.
In conclusion, GcnE plays an essential role in asexual development and is required for the expression of the master regulator of conidiation brlA and some yet-unidentified conidiation-specific genes. It is, to the best of our knowledge, so far the only nonessential histone modifier with such a role. One of the questions to be followed up is to identify the other GcnE-dependent mechanisms required for initiation of development and to elucidate the factors that recruit the SAGA complex to the promoter of brlA.
Supplementary Material
Acknowledgments
We thank Jae-Hyuk Yu, Nancy Keller, Axel Brakhage, and the Fungal Genetics Stock Center for sharing strains and Juan Luis Ribas and Cristina Vaquero (Servicio de Microscopía, Centro de Investigación Tecnología e Innovación, Universidad de Sevilla) for help with SEM. D.C. thanks the University of Sevilla for supporting his stay at BOKU–University of Natural Resources and Life Sciences, Vienna. Work was funded by grant SFB-F37-3 from the Austrian Science Fund (Fonds zur Förderung der wissenschaftlichen Forschung) and grant LS12-009 (EpiMed) of the NÖ-Forschung und Bildung Fund (to J.S.).
Footnotes
Communicating editor: A. P. Mitchell
Literature Cited
- Adams T. H., Boylan M. T., Timberlake W. E., 1988. brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell 54: 353–362. [DOI] [PubMed] [Google Scholar]
- Adams T. H., Wieser J. K., Yu J. H., 1998. Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 62: 35–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud M. B., Chibucos M. C., Costanzo M. C., Crabtree J., Inglis D. O., et al. , 2010. The Aspergillus Genome Database, a curated comparative genomics resource for gene, protein and sequence information for the Aspergillus research community. Nucleic Acids Res. 38: D420–D427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S. P., Grant P. A., 2007. The SAGA continues: expanding the cellular role of a transcriptional co-activator complex. Oncogene 26: 5329–5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayram O., Biesemann C., Krappmann S., Galland P., Braus G. H., 2008a More than a repair enzyme: Aspergillus nidulans photolyase-like CryA is a regulator of sexual development. Mol. Biol. Cell 19: 3254–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayram O., Krappmann S., Ni M., Bok J. W., Helmstaedt K., et al. , 2008b VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 320: 1504–1506. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Bok J. W., Chiang Y. M., Szewczyk E., Reyes-Dominguez Y., Davidson A. D., et al. , 2009. Chromatin-level regulation of biosynthetic gene clusters. Nat. Chem. Biol. 5: 462–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok J. W., Soukup A. A., Chadwick E., Chiang Y. M., Wang C. C., et al. , 2013. VeA and MvlA repression of the cryptic orsellinic acid gene cluster in Aspergillus nidulans involves histone 3 acetylation. Mol. Microbiol. 89: 963–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canovas D., Boyce K. J., Andrianopoulos A., 2011. The fungal type II myosin in Penicillium marneffei, MyoB, is essential for chitin deposition at nascent septation sites but not actin localization. Eukaryot. Cell 10: 302–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cove D. J., 1966. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim. Biophys. Acta 113: 51–56. [DOI] [PubMed] [Google Scholar]
- da Silva Ferreira M. E., Kress M. R., Savoldi M., Goldman M. H., Hartl A., et al. , 2006. The akuB(KU80) mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot. Cell 5: 207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon M. J., Imoto S., Nolan J., Miyano S., 2004. Open source clustering software. Bioinformatics 20: 1453–1454. [DOI] [PubMed] [Google Scholar]
- Etxebeste O., Garzia A., Espeso E. A., Ugalde U., 2010. Aspergillus nidulans asexual development: making the most of cellular modules. Trends Microbiol. 18: 569–576. [DOI] [PubMed] [Google Scholar]
- Freitag M., Williams R. L., Kothe G. O., Selker E. U., 2002. A cytosine methyltransferase homologue is essential for repeat-induced point mutation in Neurospora crassa. Proc. Natl. Acad. Sci. USA 99: 8802–8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag M., Hickey P. C., Khlafallah T. K., Read N. D., Selker E. U., 2004. HP1 is essential for DNA methylation in Neurospora. Mol. Cell 13: 427–434. [DOI] [PubMed] [Google Scholar]
- Gacek A., Strauss J., 2012. The chromatin code of fungal secondary metabolite gene clusters. Appl. Microbiol. Biotechnol. 95: 1389–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzia A., Etxebeste O., Herrero-Garcia E., Fischer R., Espeso E. A., et al. , 2009. Aspergillus nidulans FlbE is an upstream developmental activator of conidiation functionally associated with the putative transcription factor FlbB. Mol. Microbiol. 71: 172–184. [DOI] [PubMed] [Google Scholar]
- Garzia A., Etxebeste O., Herrero-Garcia E., Ugalde U., Espeso E. A., 2010. The concerted action of bZip and cMyb transcription factors FlbB and FlbD induces brlA expression and asexual development in Aspergillus nidulans. Mol. Microbiol. 75: 1314–1324. [DOI] [PubMed] [Google Scholar]
- Garzia A., Etxebeste O., Rodriguez-Romero J., Fischer R., Espeso E. A., et al. , 2013. Transcriptional changes in the transition from vegetative cells to asexual development in the model fungus Aspergillus nidulans. Eukaryot. Cell 12: 311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgakopoulos P., Lockington R. A., Kelly J. M., 2012. SAGA complex components and acetate repression in Aspergillus nidulans. G3 2: 1357–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgakopoulos P., Lockington R. A., Kelly J. M., 2013. The Spt-Ada-Gcn5 Acetyltransferase (SAGA) complex in Aspergillus nidulans. PLoS ONE 8: e65221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant P. A., Duggan L., Cote J., Roberts S. M., Brownell J. E., et al. , 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11: 1640–1650. [DOI] [PubMed] [Google Scholar]
- Grewal S. I., Jia S., 2007. Heterochromatin revisited. Nat. Rev. Genet. 8: 35–46. [DOI] [PubMed] [Google Scholar]
- Grimaldi B., Coiro P., Filetici P., Berge E., Dobosy J. R., et al. , 2006. The Neurospora crassa White Collar-1 dependent blue light response requires acetylation of histone H3 lysine 14 by NGF-1. Mol. Biol. Cell 17: 4576–4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson F. Q., Johnson T. L., 2009. Acetylation by the transcriptional coactivator Gcn5 plays a novel role in co-transcriptional spliceosome assembly. PLoS Genet. 5: e1000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson F. Q., Merkhofer E. C., Johnson T. L., 2011. Dynamic histone acetylation is critical for cotranscriptional spliceosome assembly and spliceosomal rearrangements. Proc. Natl. Acad. Sci. USA 108: 2004–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer, Ø., D. Harper, and P. D. Ryan, 2001 PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontologia Electronica 4: 1–9. [Google Scholar]
- Honda S., Lewis Z. A., Huarte M., Cho L. Y., David L. L., et al. , 2010. The DMM complex prevents spreading of DNA methylation from transposons to nearby genes in Neurospora crassa. Genes Dev. 24: 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisinga K. L., Pugh B. F., 2004. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell 13: 573–585. [DOI] [PubMed] [Google Scholar]
- Krappmann S., Sasse C., Braus G. H., 2006. Gene targeting in Aspergillus fumigatus by homologous recombination is facilitated in a nonhomologous end- joining-deficient genetic background. Eukaryot. Cell 5: 212–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krijgsheld P., Bleichrodt R., van Veluw G. J., Wang F., Muller W. H., et al. , 2013. Development in Aspergillus. Stud. Mycol. 74: 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M. H., Brownell J. E., Sobel R. E., Ranalli T. A., Cook R. G., et al. , 1996. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 383: 269–272. [DOI] [PubMed] [Google Scholar]
- Kurshakova M. M., Krasnov A. N., Kopytova D. V., Shidlovskii Y. V., Nikolenko J. V., et al. , 2007. SAGA and a novel Drosophila export complex anchor efficient transcription and mRNA export to NPC. EMBO J. 26: 4956–4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon N. J., Garzia A., Espeso E. A., Ugalde U., Yu J. H., 2010. FlbC is a putative nuclear C(2)H(2) transcription factor regulating development in Aspergillus nidulans. Mol. Microbiol. 77: 1203–1219. [DOI] [PubMed] [Google Scholar]
- Lee B. N., Adams T. H., 1994. The Aspergillus nidulans fluG gene is required for production of an extracellular developmental signal and is related to prokaryotic glutamine synthetase I. Genes Dev. 8: 641–651. [DOI] [PubMed] [Google Scholar]
- Lee I., Oh J. H., Shwab E. K., Dagenais T. R., Andes D., et al. , 2009. HdaA, a class 2 histone deacetylase of Aspergillus fumigatus, affects germination and secondary metabolite production. Fungal Genet. Biol. 46: 782–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. I., Causton H. C., Holstege F. C., Shen W. C., Hannett N., et al. , 2000. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 405: 701–704. [DOI] [PubMed] [Google Scholar]
- Li L., Zhu J., Tian J., Liu X., Feng C., 2010. A role for Gcn5 in cardiomyocyte differentiation of rat mesenchymal stem cells. Mol. Cell. Biochem. 345: 309–316. [DOI] [PubMed] [Google Scholar]
- Lin W., Srajer G., Evrard Y. A., Phan H. M., Furuta Y., et al. , 2007. Developmental potential of Gcn5(−/−) embryonic stem cells in vivo and in vitro. Dev. Dyn. 236: 1547–1557. [DOI] [PubMed] [Google Scholar]
- Martinelli S. D., Clutterbuck A. J., 1971. A quantitative survey of conidiation mutants in Aspergillus nidulans. J. Gen. Microbiol. 69: 261–268. [DOI] [PubMed] [Google Scholar]
- Medina I., Carbonell J., Pulido L., Madeira S. C., Goetz S., et al. , 2010. Babelomics: an integrative platform for the analysis of transcriptomics, proteomics and genomic data with advanced functional profiling. Nucleic Acids Res. 38: W210–W213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar C. B., Grunstein M., 2006. Genome-wide patterns of histone modifications in yeast. Nat. Rev. Mol. Cell Biol. 7: 657–666. [DOI] [PubMed] [Google Scholar]
- Mirabito P. M., Adams T. H., Timberlake W. E., 1989. Interactions of three sequentially expressed genes control temporal and spatial specificity in Aspergillus development. Cell 57: 859–868. [DOI] [PubMed] [Google Scholar]
- Munoz-Galvan S., Jimeno S., Rothstein R., Aguilera A., 2013. Histone H3K56 acetylation, Rad52, and non-DNA repair factors control double-strand break repair choice with the sister chromatid. PLoS Genet. 9: e1003237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak T., Szewczyk E., Oakley C. E., Osmani A., Ukil L., et al. , 2006. A versatile and efficient gene targeting system for Aspergillus nidulans. Genetics 172: 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninomiya Y., Suzuki K., Ishii C., Inoue H., 2004. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc. Natl. Acad. Sci. USA 101: 12248–12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nützmann H. W., Reyes-Dominguez Y., Scherlach K., Schroeckh V., Horn F., et al. , 2011. Bacteria-induced natural product formation in the fungus Aspergillus nidulans requires Saga/Ada-mediated histone acetylation. Proc. Natl. Acad. Sci. USA 108: 14282–14287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nützmann H. W., Fischer J., Scherlach K., Hertweck C., Brakhage A. A., 2013. Distinct amino acids of histone H3 control secondary metabolism in Aspergillus nidulans. Appl. Environ. Microbiol. 79: 6102–6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer J. M., Perrin R. M., Dagenais T. R., Keller N. P., 2008. H3K9 methylation regulates growth and development in Aspergillus fumigatus. Eukaryot. Cell 7: 2052–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. S., Yu J. H., 2012. Genetic control of asexual sporulation in filamentous fungi. Curr. Opin. Microbiol. 15: 669–677. [DOI] [PubMed] [Google Scholar]
- Pontecorvo G., Roper J. A., Hemmons L. M., Macdonald K. D., Bufton A. W., 1953. The genetics of Aspergillus nidulans. Adv. Genet. 5: 141–238. [DOI] [PubMed] [Google Scholar]
- Purschwitz J., Muller S., Kastner C., Schoser M., Haas H., et al. , 2008. Functional and physical interaction of blue- and red-light sensors in Aspergillus nidulans. Curr. Biol. 18: 255–259. [DOI] [PubMed] [Google Scholar]
- Reyes-Dominguez Y., Narendja F., Berger H., Gallmetzer A., Fernandez-Martin R., et al. , 2008. Nucleosome positioning and histone H3 acetylation are independent processes in the Aspergillus nidulans prnD-prnB bidirectional promoter. Eukaryot. Cell 7: 656–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Dominguez Y., Bok J. W., Berger H., Shwab E. K., Basheer A., et al. , 2010. Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans. Mol. Microbiol. 76: 1376–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Dominguez Y., Boedi S., Sulyok M., Wiesenberger G., Stoppacher N., et al. , 2012. Heterochromatin influences the secondary metabolite profile in the plant pathogen Fusarium graminearum. Fungal Genet. Biol. 49: 39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Navarro S., Fischer T., Luo M. J., Antunez O., Brettschneider S., et al. , 2004. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell 116: 75–86. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Urra A. B., Jimenez C., Nieto M. I., Rodriguez J., Hayashi H., et al. , 2012. Signaling the induction of sporulation involves the interaction of two secondary metabolites in Aspergillus nidulans. ACS Chem. Biol. 7: 599–606. [DOI] [PubMed] [Google Scholar]
- Rountree M. R., Selker E. U., 2010. DNA methylation and the formation of heterochromatin in Neurospora crassa. Heredity (Edinb) 105: 38–44. [DOI] [PubMed] [Google Scholar]
- Ruger-Herreros C., Rodriguez-Romero J., Fernandez-Barranco R., Olmedo M., Fischer R., et al. , 2011. Regulation of conidiation by light in Aspergillus nidulans. Genetics 188: 809–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinko T., Berger H., Lee W., Gallmetzer A., Pirker K., et al. , 2010. Transcriptome analysis of nitrate assimilation in Aspergillus nidulans reveals connections to nitric oxide metabolism. Mol. Microbiol. 78: 720–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeckh V., Scherlach K., Nutzmann H. W., Shelest E., Schmidt-Heck W., et al. , 2009. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 106: 14558–14563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servet C., Conde e Silva N., Zhou D. X., 2010. Histone acetyltransferase AtGCN5/HAG1 is a versatile regulator of developmental and inducible gene expression in Arabidopsis. Mol. Plant 3: 670–677. [DOI] [PubMed] [Google Scholar]
- Shwab E. K., Bok J. W., Tribus M., Galehr J., Graessle S., et al. , 2007. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot. Cell 6: 1656–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G. K., 2005. Limma: linear models for microarray data, pp. 397–420 in Computational Biology Solutions Using R and Bioconductor, edited by Gentleman V. C. R., Dudoit S., Irizarry R., Huber W. Springer, New York. [Google Scholar]
- Soukup A. A., Chiang Y. M., Bok J. W., Reyes-Dominguez Y., Oakley B. R., et al. , 2012. Overexpression of the Aspergillus nidulans histone 4 acetyltransferase EsaA increases activation of secondary metabolite production. Mol. Microbiol. 86: 314–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spedale G., Timmers H. T., Pijnappel W. W., 2012. ATAC-king the complexity of SAGA during evolution. Genes Dev. 26: 527–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss J., Reyes-Dominguez Y., 2011. Regulation of secondary metabolism by chromatin structure and epigenetic codes. Fungal Genet. Biol. 48: 62–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru H., Selker E. U., 2001. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414: 277–283. [DOI] [PubMed] [Google Scholar]
- Timberlake W. E., 1980. Developmental gene regulation in Aspergillus nidulans. Dev. Biol. 78: 497–510. [DOI] [PubMed] [Google Scholar]
- Tribus M., Bauer I., Galehr J., Rieser G., Trojer P., et al. , 2010. A novel motif in fungal class 1 histone deacetylases is essential for growth and development of Aspergillus. Mol. Biol. Cell 21: 345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R. A., Chang P. K., Dobrzyn A., Ntambi J. M., Zarnowski R., et al. , 2004. Two Delta9-stearic acid desaturases are required for Aspergillus nidulans growth and development. Fungal Genet. Biol. 41: 501–509. [DOI] [PubMed] [Google Scholar]
- Workman J. L., 2006. Nucleosome displacement in transcription. Genes Dev. 20: 2009–2017. [DOI] [PubMed] [Google Scholar]
- Yu J. H., Wieser J., Adams T. H., 1996. The Aspergillus FlbA RGS domain protein antagonizes G protein signaling to block proliferation and allow development. EMBO J. 15: 5184–5190. [PMC free article] [PubMed] [Google Scholar]
- Yu J. H., Mah J. H., Seo J. A., 2006. Growth and developmental control in the model and pathogenic aspergilli. Eukaryot. Cell 5: 1577–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.