Abstract
Tumorigenicity studies often employ outbred nude mice, in the absence of direct evidence that this mixed genetic background will negatively affect experimental outcome. Here we show that outbred nude mice carry two different alleles of Pla2g2a, a genetic modifier of intestinal tumorigenesis in mice. Here, we identify previous unreported linked polymorphisms in the promoter, noncoding and coding sequences of Pla2g2a and show that outbred nude mice from different commercial providers are heterogeneous for this polymorphic Pla2g2a allele. This heterogeneity even extends to mice obtained from a single commercial provider, which display mixed Pla2g2a genotypes. Notably, we demonstrated that the polymorphic Pla2g2a allele affects orthotopic xenograft establishment of human colon cancer cells in outbred nude mice. This finding establishes a non-cell-autonomous role for Pla2g2a in suppressing intestinal tumorigenesis. Using in vitro reporter assays and pharmacological inhibitors, we show promoter polymorphisms and nonsense-mediated RNA decay (NMD) as underlying mechanisms that lead to low Pla2g2a mRNA levels in tumor-sensitive mice. Together, this study provides mechanistic insight regarding Pla2g2a polymorphisms and demonstrates a non-cell-autonomous role for Pla2g2a in suppressing tumors. Moreover, our direct demonstration that mixed genetic backgrounds of outbred nude mice can significantly affect baseline tumorigenicity cautions against future use of outbred mice for tumor xenograft studies.
Keywords: Pla2g2a/Mom1, Apc-mouse models, nude mice, xenograft model, nonsense- mediated RNA decay
Athymic nude mice, since their first use for tumorigenicity studies in the late 1960s (Rygaard and Povlsen 1969), have served as a model in >40,000 publications. This model is used to assess the potential of tumor cells to proliferate, invade, and metastasize, as well as the efficacy of anticancer therapeutics (Singh and Ferrara 2012; Zhang et al. 2012). Nude mice are defective in T-cell-mediated immunity and, therefore, are less prone to reject human cancer cell xenografts (Rygaard and Povlsen 1969). Although originally maintained as an inbred BALB/c mouse strain, an outbred line of nude mice with increased fertility and vigor is now the mainstay for xenograft studies. It has been suggested that the genetic variation found in outbred mice better recapitulates the heterogeneity found in the natural population. While true, this same genetic variation also may confound experiments performed using small samples of outbred mice.
Genetic modifiers are powerful tools for elucidating protein and pathway interactions that influence a phenotype. With a C57Bl/6 (B6) genetic background, a mouse that harbors a germline mutation that truncates the tumor suppressor Adenomatous polyposis coli (ApcMin) develops from 20 to >100 intestinal tumors (polyps) and will die at around 20 weeks of age (Moser et al. 1992; Zeineldin and Neufeld 2013a,b). When comparing polyp numbers in ApcMin mice in different strains, an even greater variation (0.5–>300) is observed (Halberg et al. 2009). Some of this variation could be attributed to environmental factors. In addition, 18 genetic modifiers were found to contribute to these wide variations in intestinal tumor number (Kwong and Dove 2009; Zeineldin and Neufeld 2013a,b). For some of these modifiers, specific sequence variants have been defined (Kwong et al. 2007; McCart et al. 2008; Oikarinen et al. 2009; Crist et al. 2011; Nnadi et al. 2012).
The first identified and best-characterized genetic modifier of ApcMin, Mom-1 was linked to a region of chromosome 4 containing the Pla2g2a gene, which encodes a secreted phospholipase A2 enzyme (Moser et al. 1992; Dietrich et al. 1993). ApcMin mice develop fewer intestinal polyps if they are from strains that show high intestinal expression of Pla2g2a (tumor resistant) and more intestinal polyps in mouse strains that show no detectable Pla2g2a gene expression (tumor sensitive) (MacPhee et al. 1995). Overexpression of Pla2g2a with a transgene reduces the number of ApcMin-induced polyps in the sensitive B6 reference mouse strain, indicating that Pla2g2a contributes to the Mom-1 phenotype (Cormier et al. 1997). Sequencing Pla2g2a cDNA from multiple mouse strains revealed an extra base in exon 3 in tumor-sensitive but not in tumor-resistant strains (Kennedy et al. 1995; MacPhee et al. 1995). This frameshift mutation results in a premature stop and was associated with an alternatively spliced RNA (Kennedy et al. 1995; MacPhee et al. 1995). Because this nonsense mutation in tumor-sensitive strains occurs before the last exon, it was predicted to lead to nonsense-mediated RNA decay (NMD) (MacPhee et al. 1995). NMD is a conserved eukaryotic mechanism by which to reduce the burden of mutant proteins by degrading mRNAs with premature stop codons (Palacios 2013). However, NMD of the Pla2g2a transcript has not been demonstrated experimentally. In addition, whether Pla2g2a reduces intestinal tumorigenesis in a cell autonomous or non-cell-autonomous manner is not completely understood. As nude mice are maintained as outbred colonies the question remained whether they vary at Pla2g2a locus and whether Pla2g2a genotypic variation can affect tumorigenesis in human xenograft studies using nude mice.
Here we identify new polymorphisms in the promoter, exons and introns of the Pla2g2a gene and confirm the previously identified exon 3 polymorphism in Pla2g2a. We show that these linked polymorphisms are present in tumor-resistant mouse strains (Pla2g2aR) and associate with higher levels of intestinal Pla2g2a mRNA. Commercially available outbred nude mice are heterogeneous for the Pla2g2a polymorphisms, both when comparing mice from different providers and among mice from a single provider. Importantly, nude mice harboring the Pla2g2aR allele showed decreased establishment of human colon cancer cell xenografts. Finally, we also present evidence that both promoter polymorphisms and NMD participate in the mechanism underlying reduced Pla2g2a expression in tumor-sensitive strains.
Materials and Methods
Mice
The research using mice in this study was conducted in accordance with OLAW and AAALAC guidelines and was approved by the University of Kansas and the University of Kansas Medical Center Institutional Animal Care and Use Committees. We received two Apc1322T male mice as a generous gift from Dr. Ian Tomlinson, Oxford University, United Kingdom. ApcMin mice were purchased from Jackson laboratory (Bar Harbor, ME). Both Apc1322T and ApcMin mice were maintained in the University of Kansas Animal Care Unit by breeding males to wild-type C57Bl/6J females (Jackson laboratory). Mice were genotyped according to the published protocols (Pollard et al. 2009).
Screening for Pla2g2a alleles
The mouse strain that was used as the reference for the University of California, Santa Cruz (UCSC) genome sequence database came from the C57Bl/6 mouse strain (http://genome.ucsc.edu/cgi-bin/hgGateway). We refer to this reference Pla2g2a sequence as the “B6 allele.” We refer to a Pla2g2a sequence that varies from that found in C57BL/6 mice as “polymorphic.” Genomic DNA was purchased directly from Jackson laboratory, provided as gifts from researchers at The University of Kansas, or prepared from tail tissue or fecal pellets. One to two millimeters of the end of the mouse tail was incubated in 200 µl buffer [50 mM KCl, 50 mM Tris HCl (pH 8.0), 2.5 mM EDTA, 0.45% NP-40, and 0.45% Tween-20] containing 100 µg proteinase-K at 55° until tissue was completely digested. Proteinase-K was inactivated by heating at 95° for 5 min. For DNA isolation from fecal pellets (Figure 3B), each fecal pellet was collected in a 1.5-ml tube containing 1 ml phosphate buffered saline (PBS). The sample was vortexed for 2 min at the maximum speed followed by centrifugation at 700 × g for 5 min. Of the supernatant, 180 µl was taken to a new tube and 20 µl proteinase-K (20mg/ ml) was added. Proteinase-K digestion for 2 hr at 55° was followed by enzyme inactivation at 95° for 5 min. DNA was further purified using DNeasy blood and tissue kit (Qiagen) following manufacturer’s instructions.
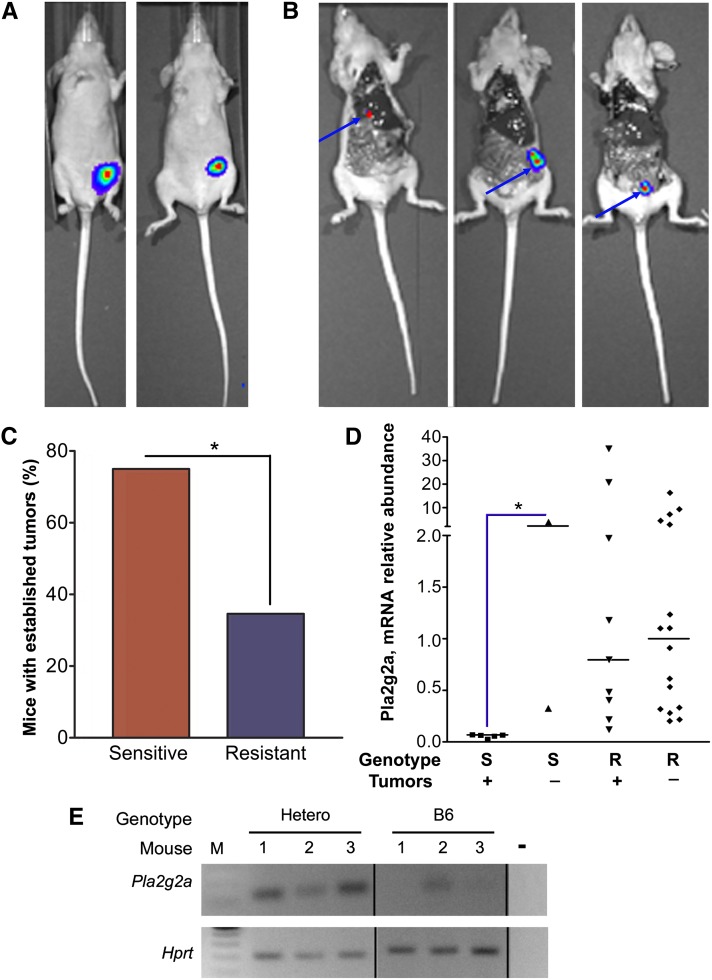
Figure 3.
The Pla2g2aR allele decreases colon cancer cell establishment in the cecum of nude mice. (A) In vivo image of luciferase-labeled HCT116 grafted in cecal wall of a sensitive (left) or resistant (right) nude mouse. (B) Ex vivo image of HCT116 luciferase-labeled cells showing hepatic metastasis (left), and local invasion (middle), both in sensitive nude mice, and local invasion (right) in a resistant nude mouse. (C) Percentage of sensitive and resistant nude mice in which HCT116 cells established tumors in the cecal wall. (*) P < 0.05 (χ2 test). (D) Quantitative RT–PCR revealed relative expression of Pla2g2a RNA in the cecum of sensitive and resistant nude mice that established or did not establish tumors in colon cancer cell orthotopic xenografts. Line represents median. (E) RT–PCR of Pla2g2a mRNA isolated from jejunal intestinal epithelial cells of B6–Pla2g2aS/S mice or mice heterozygous for the polymorphic allele (hetero–Pla2g2aR/S). The housekeeping Hprt was used as an internal control.
To distinguish the polymorphic from the B6 Pla2g2a allele, we designed a primer that is complementary to the polymorphic sequence and is different from the B6 sequence in its last two 3′ nucleotides (Figure 1B). This primer also has a mismatch mutation at the invariable nucleotide at position −4 from the 3′ end. Introducing this mismatch reduces primer annealing to the B6 Pla2g2a DNA at its 3′ region, preventing nonspecific amplification without interfering with annealing of the same primer to the polymorphic allele DNA. We used the same strategy to design a primer specific for the B6 allele that does not amplify the polymorphic allele (Table 1 shows primers sequences). PCR was performed in a 25-µl reaction containing 2 µl genomic DNA (5 µl for DNA isolated from fecal pellets), 2 mM MgCl2, 0.4 mM dNTPs (NEB), 15 pmol of the common primer (Pla2ga2F), 3 pmol of the Pla2g2aSR primer, 10 pmol of the Pla2g2aRR primer (Table 1 shows primers sequences), and 1 unit of Crimson Taq DNA polymerase (NEB). The reaction conditions were 94° for 5 min and then 30 cycles of denaturation at 94° for 30 sec, annealing at 52° for 30 sec and extension at 68° for 30 sec followed by a final extension at 68° for 3 min. PCR products were separated by electrophoresis in 2% agarose gels and detected by ethidium bromide staining and UV visualization.
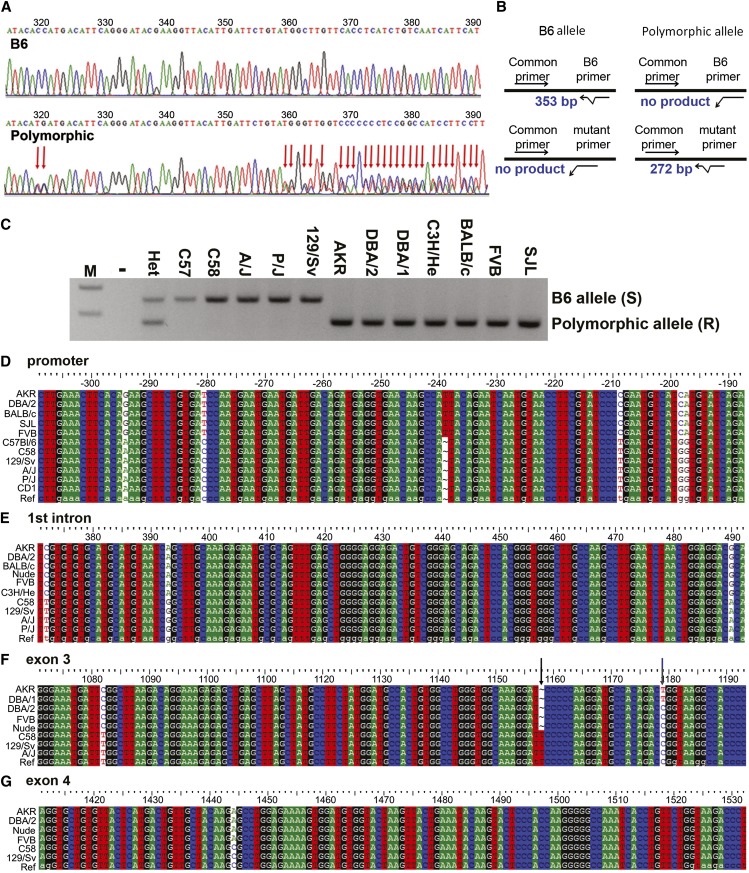
Figure 1.
Pla2g2a polymorphisms identified in ApcMin mice. (A) Chromatograph showing Pla2g2a promoter sequence for a short-lived (B6) and a long-lived (Polymorphic) Apc-mutant mouse. Polymorphisms designated by red arrows. (B) PCR-based assay for Pla2g2a polymorphisms identified in promoter sequence of long-lived (polymorphic) Apc-mutant mouse. (C) Screening different mouse strains for the Pla2g2a promoter polymorphisms using the PCR-based test. Strains AKR, DBA/2, DBA/1, C3H/He, BALB/c, FVB, and SJL carry the polymorphic promoter while strains C57, C58, A/J, P/J, and 129/Sv carry the B6 promoter. Het, heterozygous control sample from our mouse colony; −, PCR product without template; M, 100-bp DNA ladder. (D–G) Sequence results illustrate Pla2g2a polymorphisms found in different strains in the promoter area (D), first intron (E), exon 3 (F), and exon 4 (G). For exon 3 polymorphisms (E), sequence results illustrate the previously reported exon 3 nucleotide polymorphism marked by a black arrow; a newly identified R63W polymorphism in AKR and DBA/1 strains is marked by a blue arrow. Numbers represent nucleotide location, with +1 as the transcription start site.
Table 1. Primers used in this study.
| Primer name | Forward primer | Backward primer | Notes |
|---|---|---|---|
| Mom1-1 | 5′-gtaaggtggctccgtggtaa-3′ | 5′-catacccaatgccctttttg-3′ | Promoter region |
| Mom1-5′-UTR-1 | 5′-cccagtcaggagaggtttca-3′ | 5′-ggactcattccccagaattg-3′ | 5′-UTR region |
| Mom1-5′-UTR-2 | 5′-atcggcttgcaaagagaatg-3′ | 5′-ctggaaaccactgggacact-3′ | 5′-UTR region |
| Mom1-2 | 5′-ggcagttggaattcaggaaa-3′ | 5′-ttgagcctgaaaggaaatgg-3′ | |
| Mom1-3 | 5′-tcaccaccctttaccaggtc-3′ | 5′-acctcggctggctggaaac-3′ | |
| Mom1-4 | 5′-gtaaggccaccccagttctc-3′ | 5′-atcttttggccacactctgc-3′ | |
| Mom1-5 | 5′-cctaaaacagggcacacaca-3′ | 5′-agtggctgaggatgaccttg-3′ | |
| Mom1-6 | 5′-gccctctgcagtgtatgaaa-3′ | 5′tccaagttatgagaacacacacg-3′ | |
| Mom1-7 | 5′-aggccctcacaagtaaagca-3′ | 5′-cctggtttttgatggctctc-3′ | 3′-UTR region |
| Mom-2* | 5′-accatctctccagcaccaag-3′ | 5′-ggcaaaatgagacttaaatgctt-3′ | Flanks exon 3 of Atp5a |
| Mom-5* | 5′-tgttggaacgtgtttttgga-3′ | 5′-aatcgcatagatacactgtctgag-3′ | |
| Mom-7* | 5′-aacccaggactgcttccttt-3′ | 5′-ttagaaggcaggagcagagg-3′ | |
| Primers for screening for Pla2g2a promoter polymorphisms | |||
| Pla2g2aF | 5′-tgattttgaaacctctcctga-3′ | Common primer | |
| Pla2g2aSR | 5′-acttcacaaaagcttctgtaac -3′ | Wild-type- specific primer | |
| Pla2g2aRR | 5′-tcgtatccctgaatgtcttca | Polymorphism- specific primer | |
| Primers for cloning Pla2g2a promoter and 5′-UTR regions | |||
| Pla2g2a Prom | 5′-ggccgcggtaccgtgttctgcctcagctccat- 3′ | 5′-gcggcgcctaggtttcctgaattccaactgccc tt-3′ | |
| Pla2g2a 5′-UTR | 5′-ggccgccctaggaacaagacaaggccttgaacaa-3 | 5′-gcggcgccatgggctgtcagctctgtgaaggaca-3′ | |
| Primers for cloning Pla2g2a coding exons and intervening introns | |||
| Pla2 W | 5′-ggccgcccatggatgaaggtcctcctgctgctag-3′ | 5′-gcggcgtctagattgtctgatgaattgctttacttg-3′ | |
| Primers for QRT-PCR for Pla2g2a mRNA | |||
| Pla2g2a | 5′-tacaagcgcctggagaaaag-3′ | 5′-ggccttatcgcactgacaca-3′ | |
| Mouse Hprt | 5′-tgctcgagatgtcatgaagg-3′ | 5′-tatgtcccccgttgactgat-3′ | Mouse Hprt |
| Amp | 5′-ggtcctccgatcgttgtcag-3′ | 5′-cggtcgccgcatacactatt-3′ | Vector Amp |
| Human HPRT | 5′-tgacactggcaaaacaatgca-3′ | 5′-ggtccttttcaccagcaagct-3′ | |
| Human GAPDH | 5′-ccatcactgccacccagaag-3′ | 5′-agcttcccgttcagctcagg-3′ | DNA sequences |
Sequencing the Pla2g2a gene and regions of Mom-2, Mom-5, and Mom-7
The expected life spans of ApcMin and Apc1322T mice are 20 and 16 weeks, respectively (Moser et al. 1990; Pollard et al. 2009). Initial analysis of long-lived mice (all C57Bl/6 strain) included one ApcMin mouse that lived for 57 weeks and one Apc1322T mouse that lived for 37 weeks. These mice were compared to an ApcMin mouse that lived for 18 weeks and another Apc1322T mouse that became sick and was killed at 18 weeks as short-lived controls. We also included two wild-type C57Bl/6J mice purchased directly from the commercial provider (Jackson Laboratories).
Genomic DNA was amplified with overlapping primers covering the entire Pla2g2a gene, including promoter, 5′-UTR, exons, introns, and 3′-UTR (Table 1). For analysis of Mom2, Mom5, and Mom7, sequence variation-specific primers spanning published sequences were used to amplify DNA by PCR (Table 1) (Baran et al. 2007; Kwong et al. 2007; Oikarinen et al. 2009). This examination did not lead to identification of a candidate modifier. Gel-purified PCR products were sent for Sanger sequencing (ACGT Inc. or Genewiz). Sequences were compared to the C57Bl/6 mouse reference sequences at the UCSC genome browser (http://genome.ucsc.edu/cgi-bin/hgGateway). The complete Pla2g2a sequence from different strains has been uploaded in GenBank (accession nos. KF564039, KF564040, KF564041, KF564042, KF564043, KF564044, KF564045, KF564046, KF564047, KF564048, KF564049, and KF564050).
Analysis of mRNA by real-time reverse transcription–PCR
Intestinal epithelial cells were isolated from three mice with B6 Pla2g2a and three mice with polymorphic Pla2g2a alleles as described (Zeineldin et al. 2012). RNA isolation and cDNA formation from intestinal epithelial cells, HCT116 cells, or cecum of the nude mice was performed as previously described (Zeineldin et al. 2012). Briefly, 1 ml of Trizol (Invitrogen) was added to 200 µl of suspended intestinal epithelial cells. For cultured HCT116 cells, cell media was removed and cells were then scraped in 1 ml of Trizol (Invitrogen) and moved into 1.5-ml tubes. Samples were stored at −80° until use. RNA isolation and purification using Trizol (Invitrogen) was performed according to manufacturer’s instructions. For nude mouse cecal tissue, the cecum was opened and washed in PBS and a small piece (∼3 mm) was placed into a 1.5-ml tube, snap frozen in dry ice, and kept at −80° until use. Samples were homogenized in 1 ml Trizol (Invitrogen) using a hand-held homogenizer. RNA was isolated from 350 µl of the homogenized lysate using Direct-Zol RNA miniprep (Zymo reseach) following manufacturer’s protocol. The quality of the isolated RNA was assessed using 1% agarose gel electrophoresis and quantity was determined by measuring the OD260. For preparing cDNA, 1 µg total RNA was incubated for 1 hr at 42° in 1× M-MLuV enzyme buffer (NEB, Ipswich, MA) containing 1 mM dNTPs, 1 µg random hexamer primers (NEB), and 200 units of M-MLuV reverse transcriptase enzyme (NEB). The reverse transcriptase enzyme was then inactivated by heating at 95° for 5 min. Quantitative PCR was performed for mouse Pla2g2a, mouse Hprt, bacterial Amp, and Human HPRT using specific primers listed in Table 1 and using a DNA engine Opticon 2 instrument (MJ Research, Waltham, MA). The total reaction volume was 25 µl, containing 1× DyNAmo HS SYBR Green qPCR kit (Thermo), 15 pmol of each primer, and 2 µl of cDNA at a 1:5 dilution. Each reaction was performed in duplicate or triplicate. The reaction steps were initial denaturation at 95° for 15 min, followed by 40 cycles of denaturation at 94° for 20 sec, annealing at 54° for 30 sec, and extension at 72° for 30 sec. Fluorescence was measured at the end of every cycle and a melting curve was analyzed between 40° and 95° with 0.2° increments. Samples were included in the analysis only if the melting curve was a single peak at the expected temperature. For quantifying Pla2g2a mRNA in intestinal epithelial cells and ceca of nude mice, the average ΔC(t) was calculated for every sample relative to the housekeeping gene transcript Hprt. In calculating fold change of Pla2g2a mRNA from intestinal epithelial cells, ΔΔC(t) method was used relative to the C57Bl/6 mice with B6 Pla2g2a sequence. For quantification of Pla2g2a transcript in the ceca from nude mice, the relative abundance for every sample was normalized to the median value for resistant nude mice that did not establish tumors. In quantifying mouse Pla2g2a transcript in HCT116 cells, ΔC(t) was calculated relative to Amp-resistant sequence within the transfected plasmid. To correct for any variation in transfection efficiency between samples, Amp abundance was normalized to human HPRT expression and GAPDH DNA sequences. P-values were calculated using the two-tailed Mann–Whitney nonparametric test and GraphPad Prism software.
Cloning and plasmids
To test the effect of promoter polymorphisms on Pla2g2a expression, Pla2g2a-Prom(AKR):pGL3 and Pla2g2a-Prom(C58):pGL3 reporter plasmids were made by inserting Pla2g2a promoter (a 1.5-kb DNA piece upstream to Pla2g2a first codon) and 5′-UTR DNA from AKR or C58 mouse strains upstream of the Luciferase reporter gene in the pGL3 promoter vector using Pla2g2a Prom and Pla2g2a 5′-UTR primers (listed in Table 1). For testing Pla2g2a mRNA stability, all Pla2g2a coding exons and intervening introns from C58 and AKR mouse strains were used to replace the Luciferase gene in the pGL3 promoter vector using Pla2g2a whole primers (listed in Table 1). All the cloned plasmids were sequenced at ATCG and only those with no cloning-introduced mutations were included in the study.
Luciferase reporter assay
Using GeneExpresso transfection reagent following manufacturer’s instructions, 2.25 µg of either Pla2g2a-Prom(AKR):pGL3 promoter or Pla2g2a-Prom(C58):pGL3 promoter was cotransfected with 0.25 µg of Renilla luciferase (transfection control) into HCT116 cells. After 48 hr, cells were harvested and luciferase activity was assessed using a Dual Luciferase reporter assay system (Promega, Madison, WI) and a LMAXII 384 microplate reader (Molecular Devices, Sunnyvale, CA). Firefly luciferase data were normalized to Renilla luciferase. Fold change of the normalized firefly luciferase from the Pla2g2a-Prom(AKR):pGL3 promoter was calculated relative to the normalized firefly luciferase from the Pla2g2a-Prom(C58):pGL3 promoter. Data were collected from five independent experiments. P-values were calculated using the Mann–Whitney nonparametric test and GraphPad Prism software.
Testing Pla2g2a mRNA stability and NMD
To test Pla2g2 amRNA stability, HCT116 cells were transfected with 2.5 µg of Pla2g2a(AKR):pGL3 or Pla2g2a(C58):pGL3 plasmid using GeneExpresso transfection reagent following manufacturer’s instructions. After 24 hr, RNA was isolated and mouse Pla2g2a transcripts were quantified by RT–PCR using mouse-specific Pla2g2a primers. Data were collected from four independent experiments including 11 samples transfected with Pla2g2a(AKR):pGL3 and 10 samples transfected with Pla2g2a(C58):pGL3 DNA. To test NMD, transfected cells were treated with DMSO vehicle or 20 µM wortmannin for 2 hr, 100 nM cyclohexamide for 6 hr, or both before quantifying mouse Pla2g2a mRNA levels. Data were collected from three independent experiments.
Nude mice xenograft experiment
Thirty-five 6- to 8-week-old female Nu/Nu mice (provider B) were orthotopically inoculated with HCT116 cells expressing m-cherry/Luciferase. Implantation of HCT116 cells and imaging of mice were performed by investigators blind to the mouse genotypes. Briefly, mice were anesthetized with pentobarbital (50 mg/kg) and the cecum was accessed via a small incision into the peritoneal wall. With the aid of a dissecting microscope, 1 × 106 cells were injected in a volume of 60 µl into the submucosal layer of the cecum. Following survival surgery and while animals were recovering, a tail snip was collected from each animal to allow for genotyping as described above. The mice were allowed to recover and then returned to housing. The study continued for 28 days from the date of cell implantation and animals were imaged once each week using bioluminescence imaging (described below) to quantify tumor burden.
Bioluminescence imaging
Animals were injected with potassium salt of d-luciferin (150 mg/kg body weight). Following isoflurane-induced anesthesia, animals were imaged at 15 min after d-luciferin injection using a Xenogen IVIS system coupled to Living Image acquisition and analysis software version 4.0 (Perkin Elmer, Waltham, MA). Region-of-interest (ROI) boxes were drawn around the entire body (excluding tail) of the animals or around the tissue specimen for ex vivo imaging. Measurements were expressed as total flux, i.e., photons per second.
Results
Novel Pla2g2a polymorphisms identified
Genetic modifiers can alter tumorigenicity in mouse cancer models. One such modifier, Pla2g2a, has long been known to affect tumor formation in mice genetically predisposed to intestinal tumorigenesis, ApcMin. Sequence analysis of the entire Pla2g2a gene including promoter, 5′-UTR, exons, and introns revealed previously unknown sequence differences in a heterozygous state in a long-lived ApcMin mouse from our colony, but not in ApcMin mice with more typical short life spans or in wild-type C57Bl/6J mice obtained directly from a commercial provider (Figure 1A). We refer to this sequence variation as “polymorphic” when compared to the C57BL/6 reference sequence (UCSC genomic sequence database), which is referred to as B6. Many of these polymorphisms were found in the promoter region of Pla2g2a; however, some were also found in other coding and noncoding regions of the gene. The long-lived ApcMin mouse did not have alterations in other described genetic “modifiers of Min” (Mom2, Mom5, and Mom7) as determined by direct sequencing of PCR amplified genomic DNA (Baran et al. 2007; Kwong et al. 2007; Oikarinen et al. 2009).
To screen for mice with Pla2g2a polymorphisms, we developed a simple test that could accurately differentiate between the B6 allele and the polymorphic allele in a single PCR reaction. Using a common forward oligomer/primer that anneals to Pla2g2a promoter DNA in both the B6 and polymorphic alleles and two specific reverse primers that recognize either the B6 or polymorphic allele (Figure 1B), two PCR products (272 and 353 bp) are amplified.
Polymorphisms found throughout the Pla2g2a gene in different mouse strains
To unambiguously determine the sequences of the polymorphic loci, we amplified the promoter regions from each Pla2g2a allele obtained from a heterozygous mouse, introduced this DNA into a plasmid, and sequenced these plasmid DNAs. We found 26 bases that differed between the B6 and the polymorphic alleles in the Pla2g2a promoter, 5′-UTR region, and first intron. The sites of these polymorphisms relative to the transcriptional start site are; −882, −879, −874, −869, −865, −857, −851, −832, −804, −801, −692, −689, −529, −517, −479, −359, −348, −347, +48, +319, +361, +382, +479, +599, +648, and +711. As these Pla2g2a polymorphisms were not previously reported, we screened additional mouse strains using our PCR-based screening protocol and found that 7 of the 11 congenic strains also contained the polymorphisms (Figure 1C). Sequencing the Pla2g2a promoter region and 5′-UTR DNA from these strains confirmed the PCR results (Figure 1, D and E). Of note, we found that the polymorphic Pla2g2a allele was present in strains that were previously shown to be tumor resistant; AKR, DBA/1, DBA/2, BALB/c, and C3H/He (Dietrich et al. 1993; MacPhee et al. 1995; Halberg et al. 2009).
We tested whether mice with Pla2g2a promoter polymorphisms identified in our colony would also carry a previously identified Pla2g2a exon 3 alteration (absence of the T insertion found in the reference sequence from B6 mice) (Kennedy et al. 1995). Sequencing exon 3 in five mice that were heterozygous for Pla2g2a promoter polymorphisms as shown by PCR screening and promoter sequencing (Figure 1, C and D) revealed that all were heterozygous for the exon 3 T nucleotide alteration as well (not shown). In contrast, five mice that were homozygous for the B6 Pla2g2a promoter sequence were also homozygous for the corresponding B6 sequence alteration in exon 3 (a T nucleotide insertion). We conclude that the Pla2g2a allele detected in our colony and referred to as polymorphic compared to the reference B6, also has the T alteration in exon 3 that was previously reported. In addition to this alteration, we found many other “linked” polymorphisms in coding and noncoding sequences that associated with tumor-sensitive or tumor-resistant strains. These polymorphisms include a missense mutation 76 bp upstream from the original exon 3 insertion that is predicted to result in a tryptophan to arginine substitution in all tumor-sensitive strains (Figure 1F) and a similar missense mutation 21 bp downstream only in AKR and DBA/1 strains. Polymorphisms were also detected in other intron and coding exons. Figure 1G shows a polymorphic locus in exon 4. The complete Pla2g2a sequence from different strains has been uploaded in GenBank. These linked polymorphisms indicate extensive Pla2g2a sequence variation between sensitive and resistant mouse strains.
Pla2g2a polymorphisms in outbred nude mice
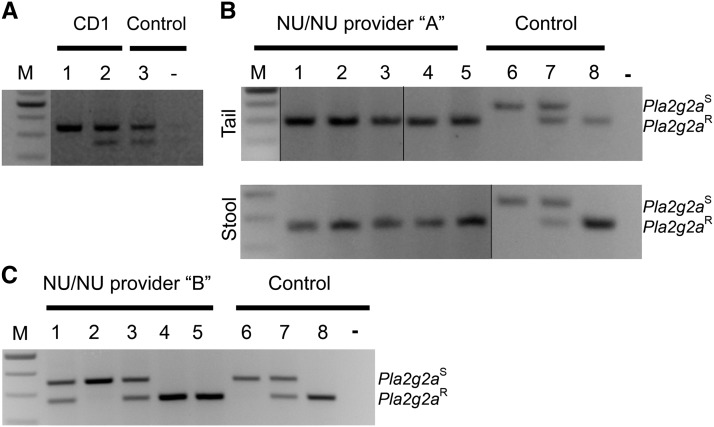
Unlike congenic inbred mouse strains that are homozygous for virtually all chromosomal loci, outbred mice maintain genetic variation in individual mice. Using our PCR-based test, we found two of the five outbred CD1 mice analyzed were heterozygous for the Pla2g2a polymorphic allele (Pla2g2aR) while the other three mice were homozygous for the B6 allele (Pla2g2aS), consistent with a previous report of the Pla2g2a exon 3 base insertion in CD1 mice (Figure 2A) (Kennedy et al. 1995). We confirmed these results by sequencing Pla2g2a from one mouse of each genotype and show the promoter sequence from a CD1 mouse in Figure 1D.
Figure 2.
Pla2g2a alleles in outbred mouse colonies. (A) PCR screening results from two CD1 mice are shown next to a control sample from a Pla2g2aR/S mouse (3). Mouse 1 is homozygous for the Pla2g2aS allele while mouse 2 is heterozygous (Pla2g2aR/S). (B) Screening nude mice from provider A using DNA from tail (top) or stool (bottom) revealed all mice to be homozygous for Pla2g2aR. (C) Screening nude mice from provider B revealed heterogeneity at the Pla2g2a locus. Mice 1 and 3 are Pla2g2aR/S, mouse 2 is Pla2g2aS/S, and mice 4 and 5 are Pla2g2aR/R. As controls for B and C, mice 6, 7, and 8 are Pla2g2aS/S, Pla2g2aR/S, and Pla2g2aR/R. −, PCR product without template. M, 100-bp DNA ladder.
Because most nude mice are maintained as outbred colonies, genetic variation is preserved among these mice. Using our PCR protocol to screen genomic DNA isolated from 103 nude mice (Nu/Nu) from two different providers, we found all 57 mice from provider A were homozygous for the Pla2g2a allele carried by resistant strains (Pla2g2aR, Figure 2B). In contrast, nude mice from provider B displayed various combinations of both alleles (Pla2g2aS and Pla2g2aR). Out of 46 mice from provider B, 10 (21.7%) were Pla2g2aS/S, 28 (60.9%) were Pla2g2aS/R, and 8 (17.4%) were Pla2g2aR/R (Figure 2C). Therefore, the frequency of the Pla2g2aS and Pla2g2aR allele in these mice was 0.543 and 0.457, respectively. DNA sequencing of the Pla2g2a promoter region and exon 3 from four Pla2g2aS/S, four Pla2g2aR/R, and three Pla2g2aR/S nude mice confirmed the presence of the B6 (Pla2g2aS) or polymorphic (Pla2g2aR) alleles, including the previously reported exon 3 insertion of T nucleotide, as predicted by PCR screening results, again indicating that these polymorphisms are linked (data not shown).
Pla2g2a affects orthotopic tumor establishment of colon cancer cells in nude mice, functioning in a non-cell-autonomous manner
Whether Pla2g2a suppression of tumorigenesis is cell autonomous or non-cell autonomous is not settled. We reasoned that, as nude mice were heterogeneous for the polymorphic Pla2g2a allele, orthotopic injection of colon cancer cells in the cecum would allow us to directly test a cell-autonomous vs. a non-cell-autonomous role for Pla2g2a in intestinal tumor suppression. In addition, this system would also allow us to test our hypothesis that Pla2g2a heterogeneity can affect the outcome of xenograft studies where human cancer cells are injected into nude mice and then tumor development and growth is monitored. To test these hypotheses, we purchased 35 female nude mice from provider B and injected luciferase-labeled HCT116 human colon cancer cells into the cecal wall of these mice. Xenograft establishment, growth, and local and distant metastasis were monitored (Figure 3A). Because the Pla2g2aR allele appears to be dominant (Dietrich et al. 1993; Cormier et al. 1997), we combined 5 Pla2g2aR/R and 22 Pla2g2aS/R mice into a single resistant group (R) with the remaining 8 Pla2g2aS/S mice forming the sensitive group (S). HCT116 cells established cecal tumors in 75% of the sensitive group mice but only 34.6% of the resistant group mice (χ2 = 4.047, two-tailed p = 0.0443, Figure 3C). Over the 4-week duration of the experiment, four of the six tumors (67%) that developed in the sensitive mice grew bigger while only 44.4% of the tumors established in resistant mice expanded (Table 2). At the end of the experiment, two tumors (33%) from mice in the sensitive group demonstrated invasion, one local, and one more distant metastasis to the liver (Figure 3B and Table 2). In the resistant group, only one tumor had local metastasis (11.1%). These results demonstrate a non-cell-autonomous role for Pla2g2a in suppression of colon tumors. Of potentially broader impact, this analysis also provides a concrete example of how a genetic modifier can significantly alter the outcome of a xenograft study performed using outbred nude mice.
Table 2. Orthotopic tumor establishment of human colon cancer cells in nude mice.
| Pla2g2a = sensitive | Pla2g2a = resistant | |
|---|---|---|
| Number | 8 | 27 |
| Mice with established tumors | 6 (75%) | 9 (34.6%) |
| Tumors growing | 4 (67%) | 4 (44.4%) |
| Liver metastasis | 1 (16.7%) | 0 |
| Local invasion | 1 (16.7%) | 1 (11.1%) |
Mice with Pla2g2a polymorphisms have more Pla2g2a mRNA in intestinal cells
The Pla2g2a allele in C57Bl/6 mice (Pla2g2aS) has been described as virtually null (MacPhee et al. 1995). However, the reduction in Pla2g2a expression was attributed to the exon 3 T insertion. Since we found previously unreported polymorphisms in Pla2g2a, with many of them in the promoter region, we were interested to measure the effects of these polymorphisms on Pla2g2a expression. We first compared Pla2g2a expression in intestinal epithelial cells from our C57BL6 mice with known Pla2g2a genotypes. We found that mice with a polymorphic allele (Pla2g2aR) had higher Pla2g2a mRNA levels in epithelial cells from jejunum, ileum, and colon (177-, 31- and 77-fold by qRT–PCR, respectively, data not shown) than mice with only the B6 allele (Pla2g2aS). Figure 3E shows RT–PCR products from jejunal RNA isolated from mice with different Pla2g2a alleles amplified with Pla2g2a- or Hprt-specific primers. Therefore, the Pla2g2a polymorphisms we initially identified in long-lived mice associate with increased expression of the gene.
Reduced Pla2g2a mRNA level also correlates with tumor sensitivity in ApcMin mice (MacPhee et al. 1995). To examine the link between establishment of orthotopic xenograft tumors and Pla2g2a expression, we performed quantitative RT–PCR using mouse-specific Pla2g2a primers and RNA isolated from ceca of nude mice. We observed wide variation in Pla2g2a mRNA levels in the cecum from nude mice, especially among the resistant group. This variation may represent heterogeneous genetic elements controlling Pla2g2a expression in these mice with mixed genetic background. Nonetheless, mice carrying the Pla2g2aR allele showed higher levels of Pla2g2a mRNA than the sensitive group mice (Figure 3D, P = 0.003). Within the sensitive group, the mice that did not establish tumors had significantly more Pla2g2a mRNA than those that established tumors (Figure 3D). The same trend of higher Pla2g2a mRNA level in mice that did not establish tumors was also seen in the resistant group, although this did not reach statistical significance.
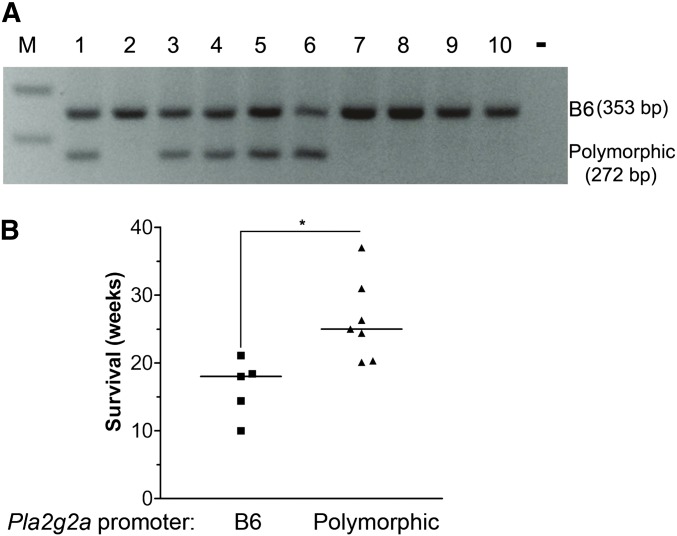
Pla2g2a polymorphisms associate with prolonged survival of Apc1322T mice
It was critical to identify all mice in our colony that had Pla2g2a polymorphisms so that they could be excluded from future studies. Using the PCR-based test to screen our mice, we found the Pla2g2a polymorphic allele in only two ApcMin mice, but in many mice with another germline Apc mutation, Apc1322T (Figure 4A). As most Apc1322T mice with the polymorphic allele were killed at different time points and many of these mice underwent different experimental conditions, compiling their intestinal tumor data were not informative. However, we found that five Apc1322T mice with the B6 Pla2g2a allele and seven Apc1322T mice heterozygous for the polymorphic allele died or were killed when they were moribund, thus providing useful survival data. While Apc1322T with the B6 Pla2g2a gene lived for 16.4 ± 1.9 weeks, which is comparable to the reported survival of Apc1322T mice (Pollard et al. 2009; Lewis et al. 2010), Apc1322T mice heterozygous for the polymorphic allele lived significantly longer, until 26.3 ± 2.3 weeks (P = 0.0075) (Figure 4B). Together, these data indicate that the Pla2g2a polymorphic allele associates with prolonged survival in Apc1322T mice.
Figure 4.
Pla2g2a polymorphisms associate with prolonged survival of Apc1322T mice. (A) Analysis of genomic DNA from 10 Apc1322T mice. Samples 1, 3, 4, 5, and 6 are from mice heterozygous for the Pla2g2a polymorphic allele, samples 2, 7, 8, 9, and 10 are from B6 mice. —, PCR product without template; M, 100-bp DNA ladder. (B) the survival of Apc1322T mice with B6 Pla2g2a status (squares) or heterozygous for the polymorphic Pla2g2a allele (triangles). Lines indicate median survival for each group. (*) P < 0.05 (two-tailed t-test).
To trace the source of the Pla2g2a polymorphic allele in our ApcMin and Apc1322T colonies, we screened the available DNA from many generations of mice. We conclude that the polymorphic Pla2g2a allele was introduced into our ApcMin and Apc1322T mice by a breeding female carrying the polymorphic allele obtained from another provider.
Promoter polymorphisms affect Pla2g2a expression
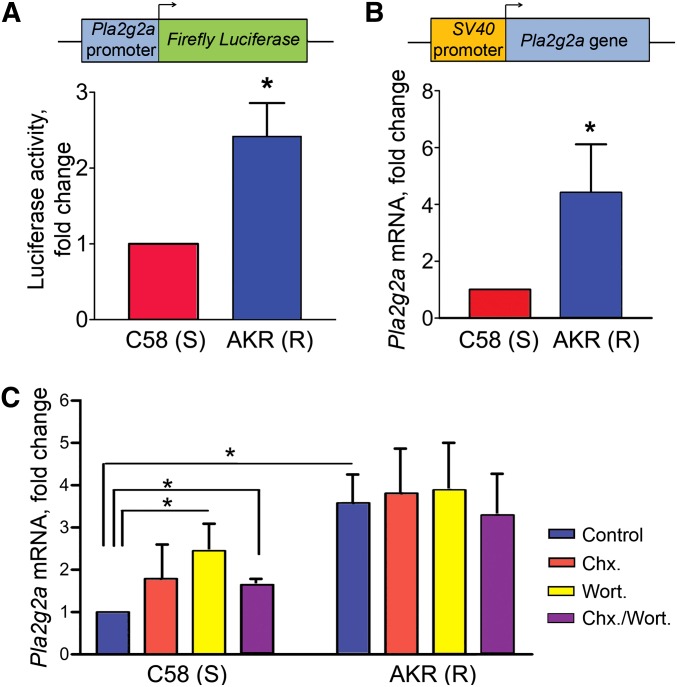
To understand the mechanisms underlying the low expression of the B6 Pla2g2a allele, we initially examined the newly identified Pla2g2a promoter polymorphisms for their contribution to gene expression. Using several programs to predict differences in transcription factor binding between the B6 and polymorphic Pla2g2a alleles, we did not identify a consistent set of transcription factors (data not shown). Nonetheless, to look for global effects of the promoter polymorphisms, we generated firefly luciferase reporter constructs driven by the Pla2g2a promoter and 5′-UTR regions, including the first intron, from the AKR strain (Pla2g2aR) or C58 strain (Pla2g2aS allele identical to that of C57Bl/6 mice) (Figure 5A). We found significantly more firefly luciferase activity in cells transfected with the AKR-derived Pla2g2a promoter and 5′-UTR reporter than in cells transfected with the C58-derived Pla2g2a promoter and 5′-UTR (Figure 5A). We conclude that the Pla2g2a promoter polymorphisms affect Pla2g2a expression, resulting in Pla2g2a RNA levels that are higher in tumor-resistant strains than in tumor-sensitive strains.
Figure 5.
Promoter polymorphisms and NMD alter Pla2g2a RNA level (A) A luciferase reporter was expressed in HCT116 cells under the control of Pla2g2a promoter DNA from the tumor-sensitive (S) C58 or the tumor-resistant (R) AKR strain. The Pla2g2a promoter from the tumor-resistant strain resulted in more luciferase than that from the sensitive strain. (*) P < 0.05 (Mann–Whitney nonparametric test). (B) qRT–PCR analysis of mouse Pla2g2a mRNA levels from the tumor-sensitive strain (C58) and the tumor-resistant strain (AKR) Pla2g2a genes each transcribed from the SV40 promoter in HCT116 cells. (*) P < 0.05 (Mann–Whitney nonparametric test). (C) NMD was inhibited using 20 µM wortmannin (Wort.) and/or 100 nM cyclohexamide (Chx.). RNA levels were quantified by qRT–PCR. Note that NMD inhibitors significantly increased C58 Pla2g2a mRNA level. (*) P < 0.05 (Mann–Whitney nonparametric test).
NMD contributes to instability of Pla2g2a transcript in tumor-resistant strains
It was previously proposed that low levels of Pla2g2a mRNA in tumor-sensitive mice were the result of NMD (MacPhee et al. 1995). This assumption, based on the known polymorphism in Pla2g2a positioned in exon 3 and thus before the last exon, has not been directly tested. To examine the potential involvement of NMD in controlling Pla2g2a levels, a second set of constructs was generated by cloning the Pla2g2a coding exons and the intervening introns from tumor-sensitive or tumor-resistant (AKR) strains after the SV40 promoter. We reasoned that as the Pla2g2a gene from each strain was controlled by the same strong promoter, any changes in transcript level would likely result from inherent RNA stability differences rather than transcription rates. Using qRT–PCR and primers specific for mouse Pla2g2a mRNA, we observed that Pla2g2a mRNA levels are significantly lower in human cells transfected with the construct containing the C58-derived Pla2g2a sequence than in those transfected with the AKR-derived Pla2g2a sequence (Figure 5B).
To further confirm a role for NMD in reduction of Pla2g2a mRNA levels in tumor-sensitive strains, we treated cultured cells with pharmacologic NMD inhibitors. Wortmannin is a PI3K inhibitor that can also inhibit the PI3K-related protein hSMG1, a component of the NMD surveillance machinery (Pal et al. 2001). Cyclohexamide inhibits eukaryotic protein synthesis and thereby also affects NMD, which requires protein synthesis for completion (Palacios 2013). When human cells transfected with the SV40-driven C58-derived Pla2g2a gene construct were treated with wortmannin alone or in combination with cyclohexamide, the mouse Pla2g2a mRNA level significantly increased relative to vehicle-treated cells (Figure 5C). In contrast, a parallel control experiment performed using the SV40-driven AKR-derived Pla2g2a gene construct revealed no change in mouse Pla2g2a transcript level in the cells treated with wortmannin, cyclohexamide, or both drugs (Figure 5C). We conclude that Pla2g2a promoter polymorphisms and NMD each contribute to Pla2g2a mRNA levels in tumor-sensitive strains.
Discussion
Genetic and environmental factors modify tumorigenicity in humans and in model organisms such as mice. Identification and characterization of genetic modifiers leads to greater appreciation for the different pathways contributing to a given phenotype (Kwong and Dove 2009). In this study, we showed that there are two distinct Pla2g2a alleles in different mouse strains: one allele is similar to that described in the reference sequence (B6 allele, Pla2g2aS) and the other is polymorphic (Pla2g2aR). The Pla2g2aR allele is different from the B6 (Pla2g2aS) allele in the promoter, coding, and noncoding sequences. We first identified this Pla2g2a polymorphic allele in long-lived ApcMin mice in our colony. Most of these polymorphisms are conserved in tumor-resistant mouse strains. We showed that Pla2g2a polymorphisms in outbred nude mice affect the orthotopic establishment of human cancer cells. In addition, we provide evidence that promoter polymorphisms affect Pla2g2a transcription and that Pla2g2a transcripts from tumor-sensitive mice undergo NMD.
Outbred mice have genetic variation that is lacking in inbred congenic mouse lines. But this same feature that provides more natural population characteristics may also confound experimental results, particularly in studies using only a small sample size (Chia et al. 2005). One of the most unexpected results from the current study was the finding that commercially available outbred nude mice are heterogeneous for Pla2g2a alleles and this heterogeneity leads to variation in the capacity of each outbred mouse to support growth of a human cancer cell xenograft (Figure 2 and Figure 3). Nude mice from two different commercial providers were not consistent in Pla2g2a genotype. While all nude mice from one provider were homozygous for the Pla2g2aR allele, both the Pla2g2aR and Pla2g2aS alleles were present in the mice from a second provider. These findings highlight the potential danger of comparing the results from studies performed using nude mice from different providers and even results from a heterogeneous mix of outbred nude mice from a single provider. For instance, nude mice in our study were shipped in seven cages with five mice per cage. As expected, there was variable distribution of Pla2g2a alleles between cages, with the frequency of Pla2g2aS/S mice in each cage ranging from 0/5 (0%) to 3/5 (60%). Results from this study unambiguously demonstrate the necessity of a large sample size in studies using outbred nude mice.
The heterogeneity of outbred nude mice is further illustrated by the varied Pla2g2a transcript levels, even in nude mice with the same Pla2g2a genotype (Figure 3D). This variation presumably results from additional differences in genetic elements that regulate gene expression. It seems likely, but remains to be tested, whether the related Pla2g2a transcript level will affect orthotopic xenografts of other colon cancer cells and other tumorigenesis studies with different cancer cells and injection sites. Nonetheless, the inability to completely monitor or control for varied expression of tumorigenesis modifiers such as Pla2g2a makes tumorigenicity studies using outbred mice problematic.
This is the first report of a genetic modifier of ApcMin that also affects the phenotype of Apc1322T mice. The Apc1322T model was developed to more closely recapitulate the longer truncated APC proteins (approximately half full length) found in human colorectal cancers (Nieuwenhuis and Vasen 2007; Pollard et al. 2009). In this study we showed that Pla2g2aR prolongs the survival of Apc1322T mice. Mom-1 was originally described as a quantitative trait locus that affects polyp multiplicity, size, and distribution in ApcMin mice. Although survival is a good indicator for polyp multiplicity, we looked retrograde at collected polyp data from our Apc1322T mice. Unfortunately, most of these mice carrying the Pla2g2aR allele were killed at different time points and were under different experimental conditions, making this analysis uninformative. A future study of Apc1322T mice with different Pla2g2a genotypes and killed at the same age would be necessary to detect any effect of Pla2g2a on polyp multiplicity, size, and distribution.
Another Apc model, the ApcΔ242 mouse, with a much shorter truncated Apc protein, showed fewer polyps in the first generation of C3H/He × C57Bl/6 mice (Crist et al. 2010). Notably, the C3H/He mouse strain has the tumor-resistant Pla2g2a allele (Figure 1C) (Markova et al. 2005). Taken together, our results in Apc1322T mice and reported data in ApcMin and ApcΔ242 mice (Dietrich et al. 1993; Crist et al. 2010) indicate that Pla2g2a polymorphisms can affect intestinal tumorigenesis in Apc-mutant mice, independent of the nature of Apc mutation.
Although Pla2g2a is responsible for most of the Mom-1 phenotype in ApcMin mice, the Pla2g2a gene is only one part of the Mom-1 locus. Another locus distal to Pla2g2a, but still within the Mom-1 locus, has a modest effect on polyp multiplicity in small intestines of ApcMin mice (Cormier et al. 2000). In this study, we examined only Pla2g2a polymorphisms in Apc1322T and nude mice and not the distal Mom-1 locus. Perhaps the distal Mom-1 locus accounts for some of the prolonged survival phenotype in Apc1322T mice. The distal Mom-1 locus might also contribute to the variation in xenograft establishment in nude mice ceca. However, the previous finding that Pla2g2a and not the distal Mom-1 locus is responsible for all the resistance phenotype in the colon (Cormier et al. 2000) argues against this possibility.
Pla2g2a does not appear to function as a universal tumor suppressor or oncogene. PLA2G2A mRNA is upregulated in lung and prostate cancer cells (Oleksowicz et al. 2012; Yu et al. 2012a). Furthermore, reducing expression of PLA2G2A by hairpin RNA reduces the proliferation of lung cancer cells, consistent with an oncogenic role for PLA2G2A (Yu et al. 2012b). On the other hand, low PLA2G2A expression in gastric tumors is associated with increased invasiveness and metastasis, with poor prognostic outcome (Ganesan et al. 2008). Additionally, mouse strains with little or no expression of Pla2g2a have increased susceptibility to intestinal tumorigenicity driven by germline Apc mutations (Dietrich et al. 1993; Crist et al. 2010), Muc2 mutations (Fijneman et al. 2008), or inflammation (Fijneman et al. 2009). Here we show that orthotopic establishment of colon cancer cells is also reduced with higher Pla2g2a expression in nude mice. These data clearly establish a protective effect for Pla2g2a against intestinal tumorigenesis in mice.
The mechanism by which the phospholipase 2A enzyme Pla2g2a decreases intestinal tumor burden in resistant strains is not completely understood. Pla2g2a is expressed in Paneth cells in the small intestine and goblet cells in the large intestine. Secreted Pla2g2a may function in a non-cell-autonomous manner by controlling bacterial flora and, in so doing, reduce inflammatory mediators that would otherwise promote tumor growth (Fijneman and Cormier 2008; Fijneman et al. 2008). Pla2g2a can also modulate signaling pathways, including Wnt signaling, potentially through regulating production of fatty acids such as arachidonic acid, a precursor for a variety of prostaglandin signaling mediators (Fijneman et al. 2009). Our orthotopic xenograft study clearly demonstrates a non-cell-autonomous role for Pla2g2a. HCT116 human colon cancer cells express little endogenous Pla2g2a (Belinsky et al. 2007), yet we found that their growth in the cecal wall of nude mice was affected in a manner that correlated with the level of cecal Pla2g2a transcript (Figure 3D). The number of polyps that develop when fetal small intestinal tissues from ApcMin mice were grafted subcutaneously was previously shown to correlate with the Mom-1 allele status (sensitive or resistant) of the isograft's donor rather than of the host mouse (Gould and Dove 1996). This result does not necessarily conflict with our cecal xenograft data, as Mom-1 could still affect intestinal tumorigencity in a non-cell-autonomous manner within isograft’s intestinal tissue microenvironment in addition to a systemic effect in the host mouse.
It was previously reported that overexpression of mouse Pla2g2a in HCT116 cells increased xenograft tumor size when these colon cancer cells were implanted subcutaneously in nude mice (Belinsky et al. 2007). This seemingly contradictory result might be explained by differences in the cell expressing Pla2g2a (implanted human cancer cells vs. surrounding mouse tissues) or the site of the xenograft implantation (subcutaneous vs. cecal wall). Moreover, in the previous study, the Pla2g2a genotype of the nude mice was not reported and this later variable would be expected to affect the experimental outcome.
In conclusion, we showed that the Pla2g2a sequence varies between tumor-resistant and tumor-sensitive mouse strains. Some of these variations in the promoter region affect Pla2g2a expression and we provide direct evidence that NMD also contributes to reducing Pla2g2a mRNA level in tumor-sensitive strains. The Pla2g2a genotype affects not only the ApcMin phenotype, but also that of Apc1322T mice. Remarkably, the Pla2g2a genotype varies in outbred nude mice from different providers and this variation affects orthotopic establishment of colon cancer cell xenografts in these mice.
Acknowledgments
We thank Dr. Ian Tomlinson, University of Oxford, UK, for generously providing us with Apc1322Tmice. We also thank Dr. Andrew L. Kung at the Dana Farber Cancer Institute for providing the FUW-Luc-mCherry-puro construct and Drs. Liang Xu, Laird Forest, Cory Berkland, Mary Lou Michaelis, Charlotte Vines, and Phillip Hardwidge from The University of Kansas and The University of Kansas Medical Center for providing mouse tissues for genotyping. We extend our gratitude to Vinit Nanavaty, Mahlet Yeshitla, William McGuinness, and Bryan Blanchat for technical support and to Staff at the Animal Care Unit at the University of Kansas for their assistance with mouse husbandry. This work was supported by RO1 CA10922 and P30 CA168524 from the National Cancer Institute, P20 RR016475 from the National Center for Research Resources, and P20 GM103418 from the National Institute of General Medical Sciences. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Communicating editor: D. W. Threadgill
Literature Cited
- Baran A. A., Silverman K. A., Zeskand J., Koratkar R., Palmer A., et al. , 2007. The modifier of Min 2 (Mom2) locus: embryonic lethality of a mutation in the Atp5a1 gene suggests a novel mechanism of polyp suppression. Genome Res. 17: 566–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belinsky G., Rajan T., Saria E., Giardina C., Rosenberg D., 2007. Expression of secretory phospholipase A2 in colon tumor cells potentiates tumor growth. Mol. Carcinog. 46: 106–116. [DOI] [PubMed] [Google Scholar]
- Chia R., Achilli F., Festing M. F., Fisher E. M., 2005. The origins and uses of mouse outbred stocks. Nat. Genet. 37: 1181–1186. [DOI] [PubMed] [Google Scholar]
- Cormier R. T., Hong K. H., Halberg R. B., Hawkins T. L., Richardson P., et al. , 1997. Secretory phospholipase Pla2g2a confers resistance to intestinal tumorigenesis. Nat. Genet. 17: 88–91. [DOI] [PubMed] [Google Scholar]
- Cormier R. T., Bilger A., Lillich A. J., Halberg R. B., Hong K. H., et al. , 2000. The Mom1AKR intestinal tumor resistance region consists of Pla2g2a and a locus distal to D4Mit64. Oncogene 19: 3182–3192. [DOI] [PubMed] [Google Scholar]
- Crist R. C., Roth J. J., Baran A. A., McEntee B. J., Siracusa L. D., et al. , 2010. The armadillo repeat domain of Apc suppresses intestinal tumorigenesis. Mamm. Genome 21: 450–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crist R. C., Roth J. J., Lisanti M. P., Siracusa L. D., Buchberg A. M., 2011. Identification of Mom12 and Mom13, two novel modifier loci of Apc (Min)-mediated intestinal tumorigenesis. Cell Cycle 10: 1092–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich W., Lander E., Smith J., Moser A., Gould K., et al. , 1993. Genetic identification of Mom-1, a major modifier locus affecting Min-induced intestinal neoplasia in the mouse. Cell 75: 631–639. [DOI] [PubMed] [Google Scholar]
- Fijneman R. J., Cormier R. T., 2008. The roles of sPLA2-IIA (Pla2g2a) in cancer of the small and large intestine. Front. Biosci. 13: 4144–4174. [DOI] [PubMed] [Google Scholar]
- Fijneman R. J., Peham J. R., van de Wiel M. A., Meijer G. A., Matise I., et al. , 2008. Expression of Pla2g2a prevents carcinogenesis in Muc2-deficient mice. Cancer Sci. 99: 2113–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fijneman R. J., Bade L. K., Peham J. R., van de Wiel M. A., van Hinsbergh V. W., et al. , 2009. Pla2g2a attenuates colon tumorigenesis in azoxymethane-treated C57BL/6 mice: expression studies reveal Pla2g2a target genes and pathways. Cell. Oncol. 31: 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan K., Ivanova T., Wu Y., Rajasegaran V., Wu J., et al. , 2008. Inhibition of gastric cancer invasion and metastasis by PLA2G2A, a novel beta-catenin/TCF target gene. Cancer Res. 68: 4277–4286. [DOI] [PubMed] [Google Scholar]
- Gould, K., and W. Dove, 1996 Action of Min and Mom1 on neoplasia in ectopic intestinal grafts. Cell Growth Differentiation 7: 1361–1368. [PubMed]
- Halberg R. B., Waggoner J., Rasmussen K., White A., Clipson L., et al. , 2009. Long-lived Min mice develop advanced intestinal cancers through a genetically conservative pathway. Cancer Res. 69: 5768–5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy B. P., Payette P., Mudgett J., Vadas P., Pruzanski W., et al. , 1995. A natural disruption of the secretory group II phospholipase A2 gene in inbred mouse strains. J. Biol. Chem. 270: 22378–22385. [DOI] [PubMed] [Google Scholar]
- Kwong L. N., Dove W. F., 2009. APC and its modifiers in colon cancer. Adv. Exp. Med. Biol. 656: 85–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong L. N., Shedlovsky A., Biehl B. S., Clipson L., Pasch C. A., et al. , 2007. Identification of Mom7, a novel modifier of Apc(Min/+) on mouse chromosome 18. Genetics 176: 1237–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A., Segditsas S., Deheragoda M., Pollard P., Jeffery R., et al. , 2010. Severe polyposis in Apc(1322T) mice is associated with submaximal Wnt signalling and increased expression of the stem cell marker Lgr5. Gut 59: 1680–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPhee M., Chepenik K. P., Liddell R. A., Nelson K. K., Siracusa L. D., et al. , 1995. The secretory phospholipase A2 gene is a candidate for the Mom1 locus, a major modifier of ApcMin-induced intestinal neoplasia. Cell 81: 957–966. [DOI] [PubMed] [Google Scholar]
- Markova M., Koratkar R. A., Silverman K. A., Sollars V. E., MacPhee-Pellini M., et al. , 2005. Diversity in secreted PLA2-IIA activity among inbred mouse strains that are resistant or susceptible to Apc Min/+ tumorigenesis. Oncogene 24: 6450–6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCart A. E., Vickaryous N. K., Silver A., 2008. Apc mice: models, modifiers and mutants. Pathol. Res. Pract. 204: 479–490. [DOI] [PubMed] [Google Scholar]
- Moser A. R., Pitot H. C., Dove W. F., 1990. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science 247: 322–324. [DOI] [PubMed] [Google Scholar]
- Moser A. R., Dove W. F., Roth K. A., Gordon J. I., 1992. The Min (multiple intestinal neoplasia) mutation: its effect on gut epithelial cell differentiation and interaction with a modifier system. J. Cell Biol. 116: 1517–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld K. L., 2009. Nuclear APC. Adv. Exp. Med. Biol. 656: 13–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwenhuis M. H., Vasen H. F., 2007. Correlations between mutation site in APC and phenotype of familial adenomatous polyposis (FAP): a review of the literature. Crit. Rev. Oncol. Hematol. 61: 153–161. [DOI] [PubMed] [Google Scholar]
- Nnadi S., Watson R., Innocent J., Gonye G., Buchberg A., et al. , 2012. Identification of five novel modifier loci of Apc(Min) harbored in the BXH14 recombinant inbred strain. Carcinogenesis 33: 1589–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikarinen S. I., Cleveland A. G., Cork K. M., Bynote K. K., Rafter J. J., et al. , 2009. Genetic mapping of Mom5, a novel modifier of Apc(Min)-induced intestinal tumorigenesis. Carcinogenesis 30: 1591–1596. [DOI] [PubMed] [Google Scholar]
- Oleksowicz L., Liu Y., Bracken R., Gaitonde K., Burke B., et al. , 2012. Secretory phospholipase A2-IIa is a target gene of the HER/HER2-elicited pathway and a potential plasma biomarker for poor prognosis of prostate cancer. Prostate 72: 1140–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal M., Ishigaki Y., Nagy E., Maquat L. E., 2001. Evidence that phosphorylation of human Upfl protein varies with intracellular location and is mediated by a wortmannin-sensitive and rapamycin-sensitive PI 3-kinase-related kinase signaling pathway. RNA 7: 5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios I. M., 2013. Nonsense-mediated mRNA decay: from mechanistic insights to impacts on human health. Brief Funct Genomics 12: 25–36. [DOI] [PubMed] [Google Scholar]
- Pollard P., Deheragoda M., Segditsas S., Lewis A., Rowan A., et al. , 2009. The Apc 1322T mouse develops severe polyposis associated with submaximal nuclear beta-catenin expression. Gastroenterology 136: 2204–2213. [DOI] [PubMed] [Google Scholar]
- Rygaard J., Povlsen C. O., 1969. Heterotransplantation of a human malignant tumour to “Nude” mice. Acta Pathol. Microbiol. Scand. 77: 758–760. [DOI] [PubMed] [Google Scholar]
- Singh M., Ferrara N., 2012. Modeling and predicting clinical efficacy for drugs targeting the tumor milieu. Nat. Biotechnol. 30: 648–657. [DOI] [PubMed] [Google Scholar]
- Yu J., Li H., Meng X., Fullerton D., Nemenoff R., et al. , 2012a Group IIa secretory phospholipase expression correlates with group IIa secretory phospholipase inhibition-mediated cell death in K-ras mutant lung cancer cells. J. Thorac. Cardiovasc. Surg. 144: 1479–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Mauchley D., Li H., Meng X., Nemenoff R., et al. , 2012b Knockdown of secretory phospholipase A2 IIa reduces lung cancer growth in vitro and in vivo. J. Thorac. Cardiovasc. Surg. 144: 1185–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeineldin M., Neufeld K. L., 2013a More than two decades of Apc modeling in rodents. Biochim. Biophys. Acta 1836: 80–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeineldin M., Neufeld K. L., 2013b Understanding phenotypic variation in rodent models with germline apc mutations. Cancer Res. 73: 2389–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeineldin M., Cunningham J., McGuinness W., Alltizer P., Cowley B., et al. , 2012. A knock-in mouse model reveals roles for nuclear Apc in cell proliferation, Wnt signal inhibition and tumor suppression. Oncogene 31: 2423–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Burnley P., Coder B., Su D.-M., 2012. Insights on FoxN1 biological significance and usages of the “nude” mouse in studies of T-lymphopoiesis. Int. J. Biol. Sci. 8: 1156–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]