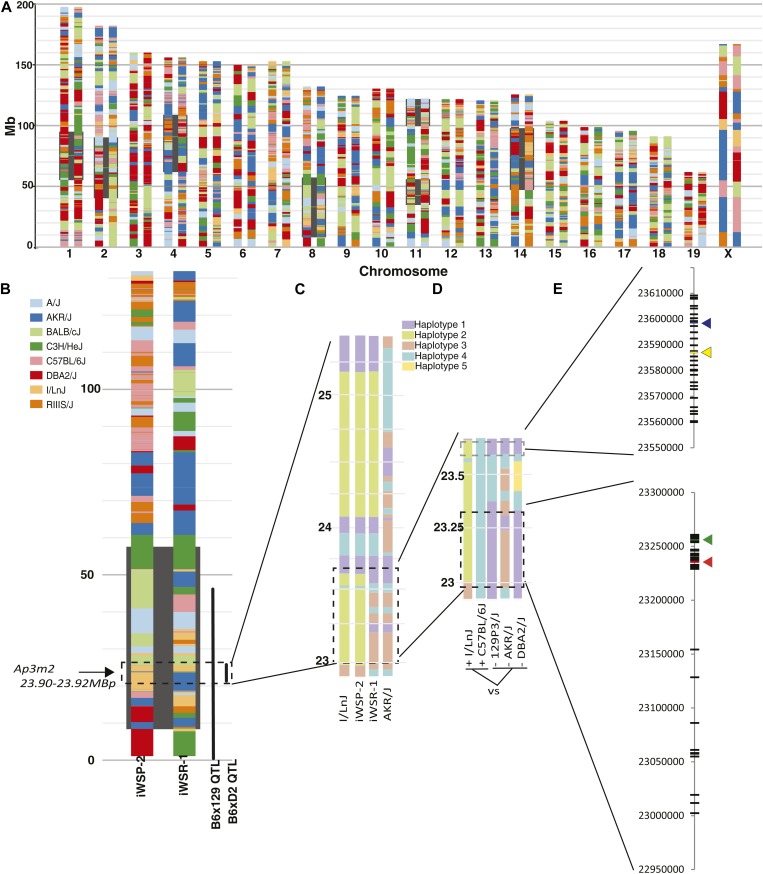
Figure 6.
Genetic dissection of shared allelic variants underlying overlapping QTL for alcohol preference and withdrawal. (A) A haplotype reconstruction of the iWSP-2 (left) and iWSR-1 (right) mouse strains based on dense genotyping of the inbred selected lines and existing genotypic data from nearest extant relatives. QTL for alcohol withdrawal seizure previously mapped in crosses of these strains are highlighted in black, including the Alcw6 locus on chromosome 8 (8.12–58.12 Mb) . The contributions of the eight HS progenitor strains are shown as determined by the Mouse Diversity Array using the SNP founder strain calls retrieved from CGDSNPdb (http://cgd.jax.org/cgdsnpdb/; Hutchins et al. 2010). (B) Enlarged reconstruction of chromosome 8 indicating location of QTL overlap of Alcw6, Ap8q (0–40.29 Mb), and BXD Ap3m2 expression QTL (22.906–25.498 Mb). The consensus QTL region on chromosome 8 is segregating I/LnJ and AKR/J haplotypes in the iWSP-2 and iWSR-1 strains. (C) Reconstruction of the iWSP-2 and iWSR-1 strain haplotypes in the region of chromosome 8 containing the consensus QTL (22.906–25.408 Mb-) from the best matching founder surrogate strains I/LnJ and AKR/J. Coloring distinguishes haplotype blocks detected in the Mouse Phylogeny Browser and does not represent genetic origin of each region. (D) Comparison of the overlapping region where iWSP-2 and iWSR-1 haplotypes differ (22.906–23.889 Mb) between reconstructed iWSP-2 and B6 (high alcohol preference, withdrawal, expression) to iWSR-1, 129, and DBA (lower alcohol preference, withdrawal, expression) haplotypes using Mouse Phylogeny Browser. Shared color blocks at a given location represent haplotype similarity across strains. (E) All genetically sufficient SNPs to account for the consensus QTL. SNPs within Ensembl regulatory features are shown with colored arrows. SNPs that contrast the strains used in the Ap8q mapping QTL (129 and B6), Alcw6 iWSP-2/iWSR-1-contributing QTL alleles (AKR/J, I/LnJ), and Ap3m2 BXD eQTL were extracted from Sanger Mouse Genomes (Keane et al. 2011; Yalcin et al. 2011; http://www.sanger.ac.uk/resources/mouse/genomes/) and are plotted in megabase coordinates on chromosome 8. iWSP-2 and iWSR-1 SNP data are publicly available at http://phenome.jax.org/ (accession no. MPD:432).