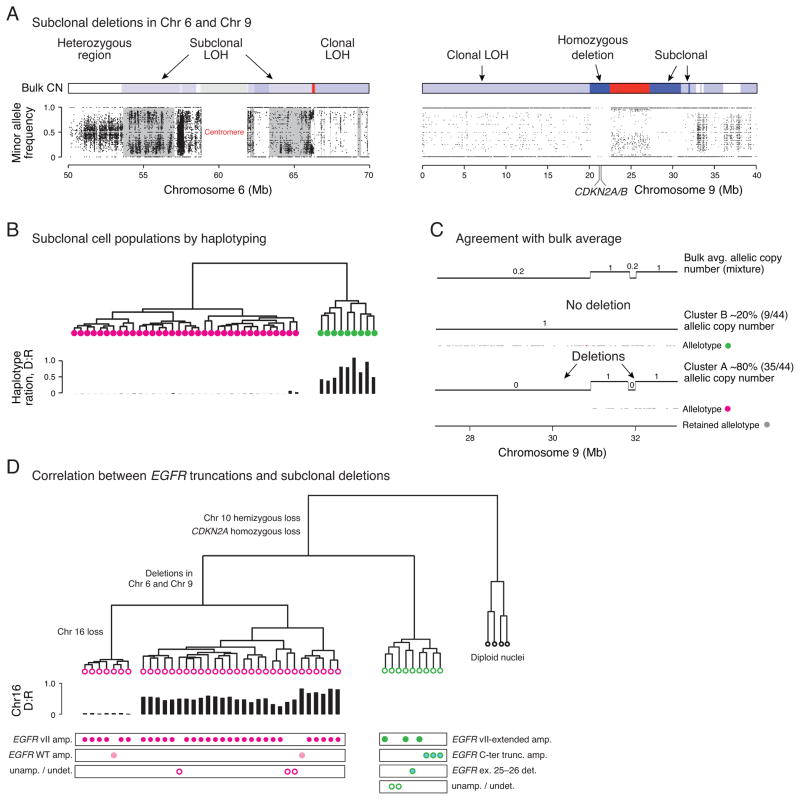
Figure 5. Subclonal deletions delineate two tumor populations harboring distinct EGFR variants.
(A) Subclonal LOH in chromosomes 6 and 9 of BT340 is reflected by incomplete segregation of SNVs from the population of single tumor nuclei. Gray masking on the left panel indicates regions of subclonal LOH flanked by regions with clonal LOH in chromosome 6. Regions of subclonal homozygous deletions in chromosome 9 have a few heterozygous sites but there are none in regions of clonal homozygous deletion. (B) Hierarchical clustering of 44 tumor nuclei from BT340 based on the single-nucleus haplotypes in regions of subclonal LOH in Chr. 6 identifies two clusters with either loss or preservation of heterozygosity. (C) Two segments in 9p21 with bulk average copy number ~0.20 (top track) are resolved as a mixture of one subclone with 80% of tumor nuclei (35/44) having deletions in both regions. Gray dots show the retained haplotype that is only present in regions that are not deleted. (D) Correlation between subclonal populations as delineated by deletions in 6p11.2, 6q12 and 9p21.1 and distinct EGFR truncations suggest that the two distinct EGFRvII truncations independently emerged after clonal segregation determined by the deletion events.